Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance - PubMed (original) (raw)
doi: 10.1371/journal.pbio.0040242.
Peter Banks, Steve Laval, Nazia A Majid, Dale Dorsett, Amer Rana, Jim Smith, Alex Bateman, Sanja Krpic, Arnd Hostert, Robert A Rollins, Hediye Erdjument-Bromage, Paul Tempst, Claire Y Benard, Siegfried Hekimi, Sarah F Newbury, Tom Strachan
Affiliations
- PMID: 16802858
- PMCID: PMC1484498
- DOI: 10.1371/journal.pbio.0040242
Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance
Vlad C Seitan et al. PLoS Biol. 2006 Jul.
Abstract
Saccharomyces cerevisiae Scc2 binds Scc4 to form an essential complex that loads cohesin onto chromosomes. The prevalence of Scc2 orthologs in eukaryotes emphasizes a conserved role in regulating sister chromatid cohesion, but homologs of Scc4 have not hitherto been identified outside certain fungi. Some metazoan orthologs of Scc2 were initially identified as developmental gene regulators, such as Drosophila Nipped-B, a regulator of cut and Ultrabithorax, and delangin, a protein mutant in Cornelia de Lange syndrome. We show that delangin and Nipped-B bind previously unstudied human and fly orthologs of Caenorhabditis elegans MAU-2, a non-axis-specific guidance factor for migrating cells and axons. PSI-BLAST shows that Scc4 is evolutionarily related to metazoan MAU-2 sequences, with the greatest homology evident in a short N-terminal domain, and protein-protein interaction studies map the site of interaction between delangin and human MAU-2 to the N-terminal regions of both proteins. Short interfering RNA knockdown of human MAU-2 in HeLa cells resulted in precocious sister chromatid separation and in impaired loading of cohesin onto chromatin, indicating that it is functionally related to Scc4, and RNAi analyses show that MAU-2 regulates chromosome segregation in C. elegans embryos. Using antisense morpholino oligonucleotides to knock down Xenopus tropicalis delangin or MAU-2 in early embryos produced similar patterns of retarded growth and developmental defects. Our data show that sister chromatid cohesion in metazoans involves the formation of a complex similar to the Scc2-Scc4 interaction in the budding yeast. The very high degree of sequence conservation between Scc4 homologs in complex metazoans is consistent with increased selection pressure to conserve additional essential functions, such as regulation of cell and axon migration during development.
Figures
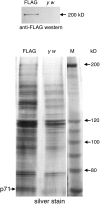
Figure 1. A 71-kDa Protein Co-Purifies with FLAG-Nipped-B by Anti-FLAG Affinity Chromatography
Nuclear extracts of_y w;_ P[Chip-FLAG-Nipped-B, w+] (FLAG) embryos and_y w_ control embryos were bound to anti-FLAG beads, washed, and eluted with FLAG peptide as described in the text. FLAG-Nipped-B fusion protein was detected in the_y w;_ P[Chip-FLAG-Nipped-B, w+] eluate, but not in the_y w_ eluate, by anti-FLAG Western blot (top panel). Other eluted proteins were detected by silver stain (bottom panel). A 71-kDa protein specific to the FLAG-Nipped-B eluate was identified by mass spectrometry as the product of the_Drosophila CG4203_ gene. It is closely related to the human MAU-2 protein. Higher molecular weight bands specific to the FLAG-Nipped-B extract contain multiple proteins whose identities could not be established unambiguously. In the bottom panel, the p71 protein and several other proteins, including the markers, became doublets when the gel was dried for photography. All appeared as single bands before drying.
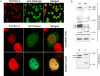
Figure 2. Human MAU-2 and Delangin Are Nuclear Proteins
(A) Confocal microscopy studies on HeLa cells. The nuclear localization of delangin is illustrated in the top panels using a FITC-labeled secondary antibody detecting monoclonal rat anti-human delangin (see Materials and Methods). Panel on the right shows merging of DNA image (left panel, red) and delangin staining (center panel, green). Expression of a GFP-human MAU-2 fusion protein revealed by confocal fluorescence microscopy of transiently transfected HeLa cells is shown in the center and bottom panels. The fusion protein appears present in both the nucleus and the cytoplasm, sometimes with a strong nuclear localization, as shown in the center panels, but in other cells cytoplasmic expression predominated. Bottom panels show retention of the GFP-human MAU-2 protein in nuclei isolated from transiently transfected HeLa cells and subsequently extracted in 0.5% Triton-X. Center and bottom panels show DNA staining with TOPRO3 (left), the GFP fluorescence signal (center), and merged images (right). (B) Nuclear location for epitopes specific to human MAU-2. Top panel: Antisera against human MAU-2 cross reacted with four major bands in whole cell extracts from HeLa cells that had been treated with the negative (-ve) control siRNA oligonucleotide. The two bands indicated by the arrows were severely reduced in intensity (by ˜90%) when the same antisera was used to blot whole cell extracts from HeLa cells that had been subjected to human MAU-2 siRNA, using the M1 siRNA oligonucleotide (see Materials and Methods). Beta actin is shown as loading control. Bottom panel: HeLa cells were separated into cytoplasmic (C) and nuclear (N) fractions (see Materials and Methods). When these fractions were immunoblotted with human MAU-2 antisera, the bands specific to human MAU-2 were almost exclusively detected in the nuclear fraction, while the background bands appeared to be cytoplasmic.
Figure 3. Co-Immunoprecipitation Studies Support Interaction between Delangin and Human MAU-2
(A) Delangin co-immunoprecipitates with a GFP-human MAU-2 fusion protein. HeLa cells were transiently transfected with a GFP-human MAU-2 fusion protein construct (see Materials and Methods) or, as a negative control, with the GFP vector on its own. Cells were fractionated into nuclear (N) and cytoplasmic (C) fractions, and aliquots of the nuclear fractions were used for immunoprecipitation with an anti-GFP antibody to generate immunoprecipitation fractions. Individual fractions were size fractionated by SDS-PAGE and immunoblotted, using antibodies against delangin (top panel) or against GFP (bottom panel) as described in Materials and Methods. The immunoprecipitation fraction from immunoprecipitating the GFP-human MAU-2 fusion protein reveals a specific band at ˜100 kDa (arrow) that is not found in the negative control sample. This is the size expected for the fusion protein (˜70 kDa for human MAU-2 and ˜ 30 kDa for GFP). The upper panel shows that the delangin antibodies identify a band of ˜ 300 kDa that is co-precipitated in the GFP-human MAU-2 immunoprecipitation sample but not in the GFP vector-only control sample. (B) Delangin siRNA. In order to validate the specificity of the monoclonal antibody to delangin, HeLa cells were transfected with siRNA oligonucleotides designed to knock down delangin using individual oligonucleotides D1–D3 (see Materials and Methods) or all three combined, D123. D2 produced the most effective delangin knockdown. The same band that was recognized by the antibody in the immunoprecipitation fraction (see A) was knocked down by ˜ 90% in cells transfected with the D2 siRNA oligonucleotide, when referenced against negative (−ve) and mock-transfected HeLa cell (MT) controls.
Figure 4. Alignment of the Conserved N-terminal Region of the Scc4/MAU-2 Family
(A) Alignment of animal and plant MAU-2 homologs referenced against the human MAU-2 sequence shown at the top. (B) Alignment of fungal Scc4 homologs referenced against the S. cerevisiae Scc4 sequence shown at the top. Numbers in parentheses at left refer to a short sequence of 16–19 amino acids that has been omitted for clarity. Protein database accession numbers at right are followed by amino acid co-ordinates for the N-terminal sequences that have been aligned. The alignment was made with the MAFFT alignment tool [ 62] and colored using the CHROMA software to highlight conserved residues [ 63].
Figure 5. Physical Interaction between Delangin and Human MAU-2 Involves N-Terminal Binding Sites
(A) Structure of delangin and human MAU-2 proteins and the positions of test constructs referred to in (B). (B) A yeast two-hybrid-based system was employed to test interaction between components of the delangin and human MAU-2 proteins as illustrated in (A) (see Materials and Methods). The system incorporated a β-galactosidase colony-lift filter assay so that positive interactions were scored by a blue color; colorless colonies signified no interaction. Full-length human MAU-2 was tested with different N-terminal components of delangin, spanning amino acids 1–277, amino acids 280–685, or amino acids 686–1170. Further mapping localized the N-terminal binding site to amino acids 1–139 of the delangin protein, which showed very strong interaction with an N-terminal human MAU-2 fragment spanning amino acids 1–115, but no binding to the remainder of the MAU-2 sequence from amino acids 116–613. A positive control showed interaction between murine p53 and SV40 large T-antigen expressed from constructs cloned in the vectors pGBK-T7 and pACT-2, respectively. A negative control demonstrated lack of interaction between the same vectors alone.
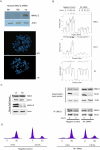
Figure 6. Human MAU-2 Regulates Sister Chromatid Cohesion and Is Required for Loading Cohesins onto Chromatin
(A) Assay for PSCS. HeLa cells were transfected with siRNA oligonucleotides (M1 and M2) designed to knock down human MAU-2 (hMAU-2). The cells were synchronized at the G2/M stage by addition of nocodazole. After 3 h, the cells in the supernatant were collected and knockdown efficiency was assayed by immunoblotting using specific antibodies against human MAU-2. Top panel, both M1 and M2 effectively knocked down human MAU-2 (˜90% and 80% knockdown, respectively) when referenced against a negative (-ve) control oligonucleotide supplied by the manufacturer; beta actin is shown as a loading control. Metaphase spreads were prepared (see Materials and Methods) from these synchronized cells and assayed for PSCS separation. Middle panel, an example of PSCS in a metaphase of HeLa cells transfected with M1. Bottom panel, a metaphase from HeLa cells transfected with a negative (-ve) control oligonucleotide (see Materials and Methods). (B) By cohesin loading assay. HeLa cells transfected with a negative control oligonucleotide or subjected to human MAU-2 knockdown using the M1 oligonucleotide were synchronised in G2/M by nocodazole treatment and then released to progress into the cell cycle. Chromatin fractions prepared from aliquots collected at 0, 1.5, 2.5, and 3.5 h were subjected to Western blotting with anti-SMC3 and anti-SCC1 antibodies to monitor loading of cohesin on the chromatin. Histone H3 was used as a loading control (lower panel). The intensity of the bands was quantified as described in Materials and Methods. (C) Left panels: Western blotting of whole Hela cell extracts that had been subjected to M1 siRNA knockdown or controls shows that the effects seen in (B) did not result from non-specific knockdown of cohesins (SMC3 and SCC1). Panels to the right: to assess the integrity of the cohesin complexes present in the supernatant, SMC3 was immunoprecipitated (see Materials and Methods) and co-immunoprecipitation of SCC1 was tested. The ratio of SMC3:SCC1 present in the immunoprecipitated samples is identical in negative control cells and cells subjected to human MAU-2 knockdown indicating that the integrity of the cohesin complexes is not affected. In the negative control there is a marked difference in the amount of immunoprecipitated SMC3 between time 0 and 3 h later, as in these cells cohesin gets loaded on the chromatin and there is fewer SMC3 available for immunoprecipitation in the supernatant (see B). In contrast, in cells subjected to human MAU-2 this difference is notably smaller, as in these cells cohesin fails to load on the chromatin to a comparable extent (see B). (D) Cohesin-loading defects observed in cells subjected to human MAU-2 knockdown are not the result of cell-cycle arrest. HeLa cells were treated as in (B) and aliquots were collected at 0 and 3.5 h. The cell-cycle profile of the cell population in each sample was analyzed by flow cytometry.
Figure 7. MAU-2 and PQN-85 Regulate Chromosome Segregation in Early C. elegans Embryos
(A) Individual RNAi knockdowns. Chromosome-segregation defects are not obvious in the progeny of_histone::GFP_ hermaphrodites injected with double-stranded_mau-2_ RNA. However, lagging anaphase chromosomes were evident in the case of_pqn-85_(RNAi) and_scc-3_(RNAi) embryos (white arrows). (B) Double RNAi knockdowns. Early_mau-2_ +pqn-85 (RNAi) embryos showed chromosome lagging (white arrow) where some ensuing cells appear to be unaffected and others have multiple and misshapen nuclei (right image). Early_mau-2_ +scc-3 (RNAi) and_pqn-85_ +scc-3 (RNAi) embryos consistently showed severe chromosome segregation defects where the DNA does not appear to move to either pole. This results in the phenotype in which all cells either have multiple nuclei or have none at all. Precise assignment of cell-cycle stages was not possible because of the severity of the chromosomal phenotype.
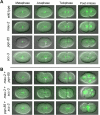
Figure 8. Delangin and MAU-2 Regulate Similar Processes in Xenopus tropicalis Embryonic Development
Antisense morpholino oligonucleotides (MO) were used to target specific mRNAs to inhibit production of X. tropicalis delangin or MAU-2 (see Materials and Methods). Embryos injected with control MO exhibit normal development (A). Embryos targeted to knock down delangin or MAU-2 both exhibit a delay in development from gastrula stages relative to control MO-injected embryos, however they look normal at this stage. By late tailbud stage (stage 28), delangin morphants (B) are severely truncated along the A-P axis and ventralized, exhibiting retarded dorsal tissue development, particularly in the neural tube and somites. Head, eye, and tail development are also defective. MAU-2 morphants (C) exhibit a very similar but less severe phenotype than is evident in delangin morphants, including shortening of the A-P axis, ventralization and defects in neural, somite, head, eye, and tail development relative to the control MO-injected embryos.
Comment in
- Ancient protein partners take on additional roles in multicellular animals.
Robinson R. Robinson R. PLoS Biol. 2006 Aug;4(8):e266. doi: 10.1371/journal.pbio.0040266. Epub 2006 Jul 4. PLoS Biol. 2006. PMID: 20076621 Free PMC article. No abstract available.
Similar articles
- Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression.
Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Watrin E, et al. Curr Biol. 2006 May 9;16(9):863-74. doi: 10.1016/j.cub.2006.03.049. Curr Biol. 2006. PMID: 16682347 - A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4.
Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP. Bernard P, et al. Curr Biol. 2006 May 9;16(9):875-81. doi: 10.1016/j.cub.2006.03.037. Curr Biol. 2006. PMID: 16682348 - Mediator recruits the cohesin loader Scc2 to RNA Pol II-transcribed genes and promotes sister chromatid cohesion.
Mattingly M, Seidel C, Muñoz S, Hao Y, Zhang Y, Wen Z, Florens L, Uhlmann F, Gerton JL. Mattingly M, et al. Curr Biol. 2022 Jul 11;32(13):2884-2896.e6. doi: 10.1016/j.cub.2022.05.019. Epub 2022 Jun 1. Curr Biol. 2022. PMID: 35654035 Free PMC article. - New insights into cohesin loading.
Litwin I, Wysocki R. Litwin I, et al. Curr Genet. 2018 Feb;64(1):53-61. doi: 10.1007/s00294-017-0723-6. Epub 2017 Jun 19. Curr Genet. 2018. PMID: 28631016 Review. - The expanding phenotypes of cohesinopathies: one ring to rule them all!
Piché J, Van Vliet PP, Pucéat M, Andelfinger G. Piché J, et al. Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review.
Cited by
- Cohesinopathies, gene expression, and chromatin organization.
Bose T, Gerton JL. Bose T, et al. J Cell Biol. 2010 Apr 19;189(2):201-10. doi: 10.1083/jcb.200912129. J Cell Biol. 2010. PMID: 20404106 Free PMC article. Review. - Condensin I folds the Caenorhabditis elegans genome.
Das M, Semple JI, Haemmerli A, Volodkina V, Scotton J, Gitchev T, Annan A, Campos J, Statzer C, Dakhovnik A, Ewald CY, Mozziconacci J, Meister P. Das M, et al. Nat Genet. 2024 Aug;56(8):1737-1749. doi: 10.1038/s41588-024-01832-5. Epub 2024 Jul 22. Nat Genet. 2024. PMID: 39039278 - Fdo1, Fkh1, Fkh2, and the Swi6-Mbp1 MBF complex regulate Mcd1 levels to impact eco1 rad61 cell growth in Saccharomyces cerevisiae.
Singh G, Skibbens RV. Singh G, et al. Genetics. 2024 Oct 7;228(2):iyae128. doi: 10.1093/genetics/iyae128. Genetics. 2024. PMID: 39110836 Free PMC article. - Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies.
Horsfield JA, Print CG, Mönnich M. Horsfield JA, et al. Front Genet. 2012 Sep 12;3:171. doi: 10.3389/fgene.2012.00171. eCollection 2012. Front Genet. 2012. PMID: 22988450 Free PMC article. - Maize Dek15 Encodes the Cohesin-Loading Complex Subunit SCC4 and Is Essential for Chromosome Segregation and Kernel Development.
He Y, Wang J, Qi W, Song R. He Y, et al. Plant Cell. 2019 Feb;31(2):465-485. doi: 10.1105/tpc.18.00921. Epub 2019 Jan 31. Plant Cell. 2019. PMID: 30705131 Free PMC article.
References
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. - PubMed
- Losada A, Hirano T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. - PubMed
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. - PubMed
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. - PubMed
- Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/C005163/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM055683/GM/NIGMS NIH HHS/United States
- R01 GM063403-01/GM/NIGMS NIH HHS/United States
- 087656/WT_/Wellcome Trust/United Kingdom
- R01 GM063403/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 GM063403-03/GM/NIGMS NIH HHS/United States
- R01 GM063403-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials