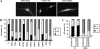Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs - PubMed (original) (raw)
Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs
Anabel Gil et al. Mol Biol Cell. 2006 Sep.
Abstract
The targeting of the tumor suppressor PTEN protein to distinct subcellular compartments is a major regulatory mechanism of PTEN function, by controlling its access to substrates and effector proteins. Here, we investigated the molecular basis and functional consequences of PTEN nuclear/cytoplasmic distribution. PTEN accumulated in the nucleus of cells treated with apoptotic stimuli. Nuclear accumulation of PTEN was enhanced by mutations targeting motifs in distinct PTEN domains, and it was dependent on an N-terminal nuclear localization domain. Coexpression of a dominant negative Ran GTPase protein blocked PTEN accumulation in the nucleus, which was also affected by coexpression of importin alpha proteins. The lipid- and protein-phosphatase activity of PTEN differentially modulated PTEN nuclear accumulation. Furthermore, catalytically active nuclear PTEN enhanced cell apoptotic responses. Our findings indicate that multiple nuclear exclusion motifs and a nuclear localization domain control PTEN nuclear localization by a Ran-dependent mechanism and suggest a proapoptotic role for PTEN in the cell nucleus.
Figures
Figure 1.
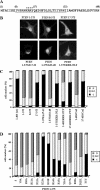
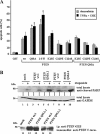
Deletions of the C-terminal tail of PTEN favor its nuclear localization. Cells were transfected with wild-type (wt) PTEN or the C-terminal PTEN truncation (1–400, 1–384, 1–375, and 1–371) or deletion (Δ350–368 and Δ370–385) mutants, as indicated. Cells were processed for immunofluorescence using the anti-PTEN 425A mAb and goat anti-mouse fluorescein-conjugated antibody (FITC-immunoglobulin). Nuclei were costained with Hoechst 33258. (A) In the top panels, representative images of the localization of each PTEN protein in U87MG cells are shown: PTEN wt, cytoplasmic (C); PTEN 1–384, 1–375, and 1–371, nuclear (N). The inset in the PTEN wt panel illustrates a cell expressing PTEN wt that localizes in both the nucleus and the cytoplasm (N/C). As expected, untransfected U87MG cells did not give positive signal for PTEN staining (unpublished data). In the bottom panels, the distribution of wt or 1–375 PTEN in HEK293 or COS-7 cells is shown. The staining corresponds to cells overexpressing recombinant PTEN. (B) Quantitation of the subcellular localization of the PTEN proteins shown in the top panels in A. Localization was classified in three different categories: N, nuclear; C, cytoplasmic; and N/C, nuclear/cytoplasmic, as illustrated in A. Data are presented as the percentage of cells in each category, and values represent the mean of at least three different experiments. SD was always <10%. (C) Schematic of the PTEN C-terminal tail (residues 350–403) illustrating the regions that contain the phosphorylation and caspase cleavage sites (residues 370–385), and the PDZ-domain binding motif (residues 401–403). (D) Quantitation of the subcellular localization of wild-type PTEN versus the 1–400, Δ350–368, and Δ370–385 PTEN mutants. Data are presented as in B.
Figure 2.
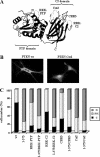
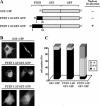
PTEN nuclear accumulation is facilitated by mutations targeting the 370–385 region of the PTEN C-terminal tail. U87MG cells were transfected with wild-type (wt) PTEN, PTEN mutations targeting the CK2 phosphorylation sites (DMA [S370A/S385A], TMA [S380A/T382A/T383A], QMA [S370A/S380A/T382A/T383A/S385A], S370A, S380A, T382A, T383A, and S385A mutations), or PTEN mutations targeting the caspase-3 cleavage sites (D384N, CM2 [D371N/D375N], and CM3 [D371N/D375N/D384N] mutations), and cells were processed for immunofluorescence as in Figure 1. (A–C) Representative images of the localization of some recombinant PTEN proteins are shown. Quantitation of the subcellular localization of the indicated PTEN proteins, using the anti-PTEN 425A mAb (B), or the anti-PTEN 421B mAb or an anti-PTEN N-terminal polyclonal antibody (C). Data are presented as in Figure 1B.
Figure 3.
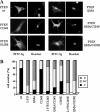
PTEN possesses multiple nuclear exclusion motifs. (A) Tertiary structure of PTEN (Lee et al., 1999), showing the surface-exposed motifs (light gray color) at the PTP and C2 domains; mutating in these motifs trigger PTEN nuclear localization. The mutations generated in these motifs that were tested for effects on PTEN localization are indicated; RKK-PTP (R161A/K163A/K164A); RRK-C2 (R233A/R234A/K237A); CBR3 (K263A/M264A/L265G/K266A/K267A/K269A); and Cα2 (K327A/N329G/K330A/K332A/R335A). (B and C) U87MG cells were transfected with wild-type (wt) PTEN or with PTEN mutations targeting the PTP and the C2 domains regions, as indicated, and cells were processed for immunofluorescence as in Figure 1. In B, representative images of the localization of the indicated PTEN proteins are shown. In C, quantitation of the subcellular localization of the indicated PTEN proteins is shown. Data are presented as in Figure 1B.
Figure 4.
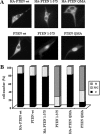
The nuclear accumulation of PTEN is affected in mutations that compromise catalytic activities. U87MG cells were transfected with wild-type (wt) PTEN, QMA (S370A/S380A/T382A/T383A/S385A), 1–375, or the catalytically inactive mutants (C124S and G129E) alone or in combination with the QMA (QMA/C124S and QMA/G129E) or the 1–375 (1–375/C124S and 1–375/G129E) mutations, and cells were processed for immunofluorescence as in Figure 1. (A) Representative images of the localization of the indicated PTEN proteins are shown (FITC-immunoglobulin). Nuclei were costained with Hoechst 33258. (B) Quantitation of the subcellular localization of the indicated PTEN proteins. Data are presented as in Figure 1B.
Figure 5.
PTEN tagged at its N-terminus with the HA epitope does not accumulate in the nucleus. U87MG cells were transfected with wild-type (wt) PTEN, QMA, or 1–375 PTEN mutants, untagged or tagged at their N-termini with the HA epitope (HA-PTEN), and the cells were assessed for PTEN localization as in Figure 1. (A) Representative images of wild-type and mutant PTEN proteins are shown. (B) Quantitation of the subcellular localization of the PTEN proteins shown in A. Data are presented as in Figure 1B.
Figure 6.
Nuclear accumulation of PTEN depends on its N-terminal domain. (A) Amino acid sequence of the N-terminal region of PTEN. Numbers in brackets correspond to PTEN amino acid numbering. The region required for PTEN nuclear accumulation is underlined. Asterisks indicate residues essential for PTEN nuclear accumulation. (B–D) U87MG cells were transfected with wild-type (wt) PTEN or with PTEN mutations, as indicated, and cells were processed for immunofluorescence as in Figure 1. The compound mutations analyzed include KRR-NLS (K13A/R14A/R15A), DLDL (D22A/L23A/D24A/L25A), and TYY (T26A/Y27A/Y29A). RKK-PTP and Cα2 mutations are as in Figure 3. In B, representative images of the localization of some PTEN proteins are shown. (C and D) Quantitation of the subcellular localization of the indicated PTEN proteins. Data are presented as in Figure 1B.
Figure 7.
The N-terminus of PTEN contains a functional nuclear localization domain. (A) Schematic representation of the GST-GFP and PTEN/GST-GFP chimeric fusion proteins used. The black boxes denote the N-terminal region of PTEN. As a summary of the results shown in B and C, the nuclear localization of the chimeras is indicated at the right. (B and C) U87MG cells were transfected with GST-GFP or with GST-GFP containing the first 16 or 32 residues of PTEN (PTEN 1–16/GST-GFP, or PTEN 1–32/GST-GFP), as indicated, and protein localization was determined by immunofluorescence as in Figure 1. In B, representative images of the distribution of the chimeric fusion proteins are shown. In C, the quantitation of the subcellular localization of the chimeras is shown. Data are presented as in Figure 1B.
Figure 8.
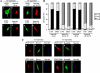
PTEN nuclear accumulation is affected by importin α proteins and depends on Ran-GTPase activity. (A and B) U87MG cells were transfected with GST-importin α1 or α5 (wild-type [wt] or Δ1–70 mutations) together with the PTEN 1–375 mutant, and cells were processed for double immunofluorescence. PTEN was detected with the anti-PTEN 425A mAb and goat anti-mouse fluorescein-conjugated secondary antibody (green; a, c, e, and g). GST-importin proteins were detected with a rabbit polyclonal anti-GST antibody and goat anti-rabbit rhodamine-conjugated secondary antibody (red; b, d, f, and h). In A, representative images of the localization of importin and PTEN 1–375 proteins are shown. In B, quantitation of the subcellular localization of PTEN 1–375 and PTEN QMA coexpressed with the importin proteins is shown. Data are presented as in Figure 1B. (C) U87MG cells were transfected with Flag-Ran (wild-type [wt] or RanQ69L mutation) together with PTEN 1–375 or QMA mutants, and cells were processed for double immunofluorescence. PTEN was detected with rabbit anti-PTEN N-terminal polyclonal antibody and goat anti-rabbit rhodamine-conjugated secondary antibody (red; a, d, g, and j). Flag-Ran proteins were detected with a mouse anti-Flag mAb and goat anti-mouse fluorescein-conjugated secondary antibody (green; b, e, h, and k). Nuclei were costained with Hoechst 33258 (c, f, i, and l). Representative images of the localization of Ran and PTEN proteins are shown.
Figure 9.
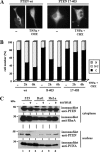
Nuclear accumulation of PTEN is augmented after treatment with agents that induce apoptosis. (A and B) U87MG cells, transfected with wild-type (wt) PTEN, 8–403 or 17–403 PTEN mutants were left untreated (−) or were treated with recombinant TNF-α plus cycloheximide (CHX) for 2 or 6 h, and then cells were processed for immunofluorescence as in Figure 1. In A, representative images of the localization of wild-type or PTEN 17–403 proteins, in untreated (−) or TNF-α plus CHX (6 h) treated cells, are shown. In B, quantitation of the subcellular localization of the indicated PTEN proteins, is shown. Data are presented as in Figure 1B. (C) 3T3, Rat1, and HeLa cells were left untreated (−) or were treated with 0.5 M sorbitol for 30 min (+). After lysis, cells were fractionated in cytoplasmic (top panels) and nuclear (bottom panels) fractions, and equal amounts of proteins were analyzed by immunoblot for endogenous PTEN, RhoA or PCNA (used as cytoplasmic and nuclear markers, respectively), using specific antibodies.
Figure 10.
Nuclear accumulation of PTEN enhances apoptosis. (A) U87MG cells, transfected with GST, wild-type (wt) PTEN, QMA (S370A, S380A, T382A, T383A, and S385A) or 1–375 PTEN mutants, or with K13E or the catalytically inactive PTEN mutants C124S or G129E (alone or in combination with the QMA mutation), were cultured in medium without serum for 24 h, followed by treatment with recombinant TNF-α plus cycloheximide (CHX) for 2 h or with doxorubicin for 24 h, as indicated. Cells were processed for immunofluorescence and stained with the anti-PTEN 425A mAb or with an anti-GST polyclonal antibody, and goat anti-mouse or anti-rabbit fluorescein conjugated secondary antibodies. Nuclei were stained with Hoechst 33258, and the percentage of apoptotic-stained cells (cells with condensed nuclei) is shown. Values represent the mean ± SD of at least two separate experiments. Significant increases with respect to wild-type PTEN are denoted with asterisks (p < 0.05). (B) HEK293 cell clones containing empty pIRES (mock) or expressing wild-type PTEN (clones wt.1 and wt.2) or PTEN QMA (clones QMA.1 and QMA.2) were left untreated (−) or were treated with 50 μM etoposide for 24 h (+). Cell were lysed, and PARP was detected from the total lysates (100 μg) using an anti-cleaved PARP-specific antibody. The amount of GAPDH, detected with an anti-GAPDH mAb, is shown as a control for protein loading. In the bottom panel (lanes 11–15), the expression of recombinant PTEN in the clones is shown. Proteins were immunoprecipitated (i.p.) with the anti-PTEN 421B mAb, followed by immunoblot with a polyclonal anti-PTEN C-terminal antibody.
Figure 11.
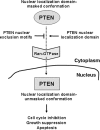
The regulation of PTEN localization. The model shown is based on results reported here and on the findings of others (Vazquez et al., 2001; Ginn-Pease and Eng, 2003; Liu, F. et al., 2005; Chung and Eng, 2005). In this model, changes in the conformation of PTEN following receipt of apoptotic or cell growth inhibitory signals would unmask the N-terminal nuclear localization domain and trigger PTEN nuclear entry by a Ran-dependent mechanism.
Similar articles
- A functional dissection of PTEN N-terminus: implications in PTEN subcellular targeting and tumor suppressor activity.
Gil A, Rodríguez-Escudero I, Stumpf M, Molina M, Cid VJ, Pulido R. Gil A, et al. PLoS One. 2015 Apr 15;10(4):e0119287. doi: 10.1371/journal.pone.0119287. eCollection 2015. PLoS One. 2015. PMID: 25875300 Free PMC article. - Cytoplasmic p27Kip1 counteracts the pro-apoptotic function of the open conformation of PTEN by retention and destabilization of PTEN outside of the nucleus.
Andrés-Pons A, Gil A, Oliver MD, Sotelo NS, Pulido R. Andrés-Pons A, et al. Cell Signal. 2012 Feb;24(2):577-587. doi: 10.1016/j.cellsig.2011.10.012. Epub 2011 Oct 20. Cell Signal. 2012. PMID: 22036806 - Identification of a nuclear localization signal in the polo box domain of Plk1.
Lee MS, Huang YH, Huang SP, Lin RI, Wu SF, Li C. Lee MS, et al. Biochim Biophys Acta. 2009 Oct;1793(10):1571-8. doi: 10.1016/j.bbamcr.2009.07.005. Epub 2009 Jul 23. Biochim Biophys Acta. 2009. PMID: 19631697 - The nuclear affairs of PTEN.
Planchon SM, Waite KA, Eng C. Planchon SM, et al. J Cell Sci. 2008 Feb 1;121(Pt 3):249-53. doi: 10.1242/jcs.022459. J Cell Sci. 2008. PMID: 18216329 Review. - Nucleocytoplasmic protein transport and recycling of Ran.
Yoneda Y, Hieda M, Nagoshi E, Miyamoto Y. Yoneda Y, et al. Cell Struct Funct. 1999 Dec;24(6):425-33. doi: 10.1247/csf.24.425. Cell Struct Funct. 1999. PMID: 10698256 Review.
Cited by
- Thioredoxin and thioredoxin target proteins: from molecular mechanisms to functional significance.
Lee S, Kim SM, Lee RT. Lee S, et al. Antioxid Redox Signal. 2013 Apr 1;18(10):1165-207. doi: 10.1089/ars.2011.4322. Epub 2012 Jun 26. Antioxid Redox Signal. 2013. PMID: 22607099 Free PMC article. Review. - Precise Immunodetection of PTEN Protein in Human Neoplasia.
Pulido R, Mingo J, Gaafar A, Nunes-Xavier CE, Luna S, Torices L, Angulo JC, López JI. Pulido R, et al. Cold Spring Harb Perspect Med. 2019 Dec 2;9(12):a036293. doi: 10.1101/cshperspect.a036293. Cold Spring Harb Perspect Med. 2019. PMID: 31501265 Free PMC article. Review. - AMPK/TSC2/mTOR-signaling intermediates are not necessary for LKB1-mediated nuclear retention of PTEN tumor suppressor.
Liu JL, Mao Z, Gallick GE, Yung WK. Liu JL, et al. Neuro Oncol. 2011 Feb;13(2):184-94. doi: 10.1093/neuonc/noq163. Epub 2010 Dec 1. Neuro Oncol. 2011. PMID: 21123367 Free PMC article. - Potentiation by Protein Synthesis Inducers of Translational Readthrough of Pathogenic Premature Termination Codons in PTEN Isoforms.
Torices L, Nunes-Xavier CE, Pulido R. Torices L, et al. Cancers (Basel). 2024 Aug 13;16(16):2836. doi: 10.3390/cancers16162836. Cancers (Basel). 2024. PMID: 39199607 Free PMC article. - Exosome-based strategies for diagnosis and therapy of glioma cancer.
Karami Fath M, Azami J, Masoudi A, Mosaddeghi Heris R, Rahmani E, Alavi F, Alagheband Bahrami A, Payandeh Z, Khalesi B, Dadkhah M, Pourzardosht N, Tarhriz V. Karami Fath M, et al. Cancer Cell Int. 2022 Aug 21;22(1):262. doi: 10.1186/s12935-022-02642-7. Cancer Cell Int. 2022. PMID: 35989351 Free PMC article. Review.
References
- Al-Khouri A. M., Ma Y., Togo S. H., Williams S., Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase PTEN by casein kinases and glycogen synthase kinase 3β. J. Biol. Chem. 2005;280:35195–35202. - PubMed
- Andrés-Pons A., Valiente M., Torres J., Gil A., Roglá I., Ripoll F., Cervera J., Pulido R. Functional definition of relevant epitopes on the tumor suppressor PTEN protein. Cancer Lett. 2005;223:303–312. - PubMed
- Aniento F., Roche E., Cuervo A. M., Knecht E. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 1993;268:10463–10470. - PubMed
- Birle D., Bottini N., Williams S., Huynh H., deBelle I., Adamson E., Mustelin T. Negative feedback regulation of the tumor suppressor PTEN by phosphoinositide–induced serine phosphorylation. J. Immunol. 2002;169:286–291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous