Whence cometh the allosterome? - PubMed (original) (raw)
Whence cometh the allosterome?
Janet E Lindsley et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Regulation of cellular functions can be accomplished by many mechanisms, including transcriptional regulation, alternative splicing, translational regulation, phosphorylation and other posttranslational covalent modifications, degradation, localization, protein-protein interactions, and small-molecule allosteric effectors. Largely because of advances in the techniques of molecular biology in the past few decades, our knowledge of regulation by most of these mechanisms has expanded enormously. Regulation by small-molecule, allosteric interactions is an exception. Many of the best-known allosteric regulators were discovered decades ago, when we knew little about all of these other forms of regulation. Allostery is the most direct, rapid, and efficient regulatory mechanism to sense changes in the concentration of small molecules and alter cellular responses to maintain homeostasis. In this perspective, we present the argument that allosteric regulation is underappreciated in the systems biology world and that many allosteric effectors remain to be discovered.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
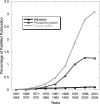
Fig. 1.
Percent of papers listed in PubMed (
www.ncbi.nlm.nih.gov/entrez/query.fcgi
) describing transcription, phosphorylation, and allostery during the past 4 decades. Titles and abstracts were searched for the following terms: “alloster*,” “protein kinase” OR “tyrosine kinase” OR “ser/thr kinase” OR “phosphorylase kinase” OR “phosphoprotein” OR “protein phosphorylation,” and “transcription*.” The number of papers identified was divided by the total number of papers published during the time interval.
Similar articles
- [G protein-coupled receptors: allosteric regulators of cell metabolism].
Galzi JL, Ilien B. Galzi JL, et al. Med Sci (Paris). 2012 Oct;28(10):852-7. doi: 10.1051/medsci/20122810013. Epub 2012 Oct 12. Med Sci (Paris). 2012. PMID: 23067416 Review. French. - Allosteric small-molecule kinase inhibitors.
Wu P, Clausen MH, Nielsen TE. Wu P, et al. Pharmacol Ther. 2015 Dec;156:59-68. doi: 10.1016/j.pharmthera.2015.10.002. Epub 2015 Oct 24. Pharmacol Ther. 2015. PMID: 26478442 Review. - Unraveling allosteric landscapes of allosterome with ASD.
Liu X, Lu S, Song K, Shen Q, Ni D, Li Q, He X, Zhang H, Wang Q, Chen Y, Li X, Wu J, Sheng C, Chen G, Liu Y, Lu X, Zhang J. Liu X, et al. Nucleic Acids Res. 2020 Jan 8;48(D1):D394-D401. doi: 10.1093/nar/gkz958. Nucleic Acids Res. 2020. PMID: 31665428 Free PMC article. - Revealing the allosterome: systematic identification of metabolite-protein interactions.
Orsak T, Smith TL, Eckert D, Lindsley JE, Borges CR, Rutter J. Orsak T, et al. Biochemistry. 2012 Jan 10;51(1):225-32. doi: 10.1021/bi201313s. Epub 2011 Dec 9. Biochemistry. 2012. PMID: 22122470 - Engineering allosteric regulation into biological catalysts.
Fastrez J. Fastrez J. Chembiochem. 2009 Dec 14;10(18):2824-35. doi: 10.1002/cbic.200900590. Chembiochem. 2009. PMID: 19937897 Review.
Cited by
- Modeling the Contribution of Allosteric Regulation for Flux Control in the Central Carbon Metabolism of E. coli.
Machado D, Herrgård MJ, Rocha I. Machado D, et al. Front Bioeng Biotechnol. 2015 Oct 8;3:154. doi: 10.3389/fbioe.2015.00154. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 26501058 Free PMC article. - Unwinding the Wnt action of casein kinase 1.
Yim DG, Virshup DM. Yim DG, et al. Cell Res. 2013 Jun;23(6):737-8. doi: 10.1038/cr.2013.51. Epub 2013 Apr 9. Cell Res. 2013. PMID: 23567556 Free PMC article. - Deciphering regulatory architectures from synthetic single-cell expression patterns.
Pan RW, Röschinger T, Faizi K, Garcia H, Phillips R. Pan RW, et al. bioRxiv [Preprint]. 2024 Jun 5:2024.01.28.577658. doi: 10.1101/2024.01.28.577658. bioRxiv. 2024. PMID: 38352569 Free PMC article. Updated. Preprint. - Glycolytic flux-signaling controls mouse embryo mesoderm development.
Miyazawa H, Snaebjornsson MT, Prior N, Kafkia E, Hammarén HM, Tsuchida-Straeten N, Patil KR, Beck M, Aulehla A. Miyazawa H, et al. Elife. 2022 Dec 5;11:e83299. doi: 10.7554/eLife.83299. Elife. 2022. PMID: 36469462 Free PMC article. - ASD: a comprehensive database of allosteric proteins and modulators.
Huang Z, Zhu L, Cao Y, Wu G, Liu X, Chen Y, Wang Q, Shi T, Zhao Y, Wang Y, Li W, Li Y, Chen H, Chen G, Zhang J. Huang Z, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D663-9. doi: 10.1093/nar/gkq1022. Epub 2010 Nov 4. Nucleic Acids Res. 2011. PMID: 21051350 Free PMC article.
References
- Bohr C. Zentr. Physiol. 1903;17:682.
- Cori G., Colowick S., Cori C. J. Biol. Chem. 1938;123:381–389.
- Crick F. Symp. Soc. Exp. Biol. 1958;XII:138. - PubMed
- Novick A., Szilard L. In: Dynamics of Growth Processes. Boell E. J., editor. Princeton: Princeton Univ. Press; 1954. pp. 21–32.
- Pardee A. B., Yates R. A. J. Biol. Chem. 1956;221:757–770. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous