Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique - PubMed (original) (raw)
Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique
Mingming Wu et al. Appl Environ Microbiol. 2006 Jul.
Abstract
An ability to monitor bacterial locomotion and collective dynamics is crucial to our understanding of a number of well-characterized phenotypes including biofilm formation, chemotaxis, and virulence. Here, we report the tracking of multiple swimming Escherichia coli cells in three spatial dimensions and at single-cell resolution using a novel three-dimensional (3D) defocused particle tracking (DPT) method. The 3D trajectories were generated for wild-type Escherichia coli strain RP437 as well as for isogenic derivatives that display smooth swimming due to a cheA deletion (strain RP9535) or incessant tumbling behavior due to a cheZ deletion (strain RP1616). The 3D DPT method successfully differentiated these three modes of locomotion and allowed direct calculation of the diffusion coefficient for each strain. As expected, we found that the smooth swimmer diffused more readily than the wild type, and both the smooth swimmer and the wild-type cells exhibited diffusion coefficients that were at least two orders of magnitude larger than that of the tumbler. Finally, we found that the diffusion coefficient increased with increasing cell density, a phenomenon that can be attributed to the hydrodynamic disturbances caused by neighboring bacteria.
Figures
FIG. 1.
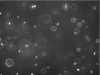
Microscope image of swimming E. coli strain RP437 (wild type for motility and chemotaxis). The image size is 449 μm by 335 μm. The bright oblong spots depict cells that are in the plane of focus. The rings depict cells that are out of the plane of focus. The ring size is directly proportional to the distance of the cell to the focal plane and is used to obtain the z dimension.
FIG. 2.
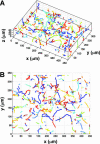
Swimming trajectories of wild-type RP437 derived from a sequence of images using ImagePro Plus (Media Cybernetics, Inc.) image processing software. The typical time between consecutive images was 150 ms, and each time sequence consisted of 300 images. Using an in-house software package, we determined the locations and the ring size for each bacterium shown in Fig. 1. This information was then translated into the coordinates (x, y, and z) for each bacterial cell in the image, and the process was repeated for all the images in a time series. The 3D trajectories of each bacterial cell generated using the nearest-neighbor method (A) and the projection of the 3D trajectory to the _x_-y plane (B). Different colors represent different trajectories.
FIG. 3.
Swimming trajectories of smooth swimmers (RP9535) and tumblers (RP1616). 3D trajectories (A) and 2D projection (B) for strain RP9535. 3D trajectories (C) and 2D projection (D) for strain RP1616. Different colors represent different trajectories.
FIG. 4.
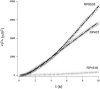
The population average of the square distance to the origin as a function of time (t) for the wild-type RP437 strain (circle) and its isogenic derivatives RP9535 (triangle) and RP1616 (diamond) at a cell concentration of 107 cells/ml. The solid lines for RP9535 and RP437 are fits to equation 1 (see text). The solid line for RP1616 is a fit to a linear function. The diffusion coefficients D and the characteristic times τ were extracted from the fits. For RP437, D = 53.2 μm2/s and τ = 1.8 s; for RP9535, D = 458.0 μm2/s and τ = 6.6 s; for RP1616, D = 2.0 μm2/s.
FIG. 5.
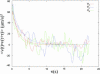
Velocity auto-correlation of wild-type E. coli strain RP437 for velocity components along the x, y, and z directions (red, vx; blue, vy; green, vz). The solid lines depict fits to an exponential decay function.
Similar articles
- Single particle tracking of correlated bacterial dynamics.
Soni GV, Ali BM, Hatwalne Y, Shivashankar GV. Soni GV, et al. Biophys J. 2003 Apr;84(4):2634-7. doi: 10.1016/s0006-3495(03)75068-1. Biophys J. 2003. PMID: 12668471 Free PMC article. - Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation.
Hyakutake A, Homma M, Austin MJ, Boin MA, Häse CC, Kawagishi I. Hyakutake A, et al. J Bacteriol. 2005 Dec;187(24):8403-10. doi: 10.1128/JB.187.24.8403-8410.2005. J Bacteriol. 2005. PMID: 16321945 Free PMC article. - TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli.
Stewart RC, VanBruggen R, Ellefson DD, Wolfe AJ. Stewart RC, et al. Biochemistry. 1998 Sep 1;37(35):12269-79. doi: 10.1021/bi980970n. Biochemistry. 1998. PMID: 9724541 - Information processing in bacterial chemotaxis.
Stock JB, Levit MN, Wolanin PM. Stock JB, et al. Sci STKE. 2002 May 14;2002(132):pe25. doi: 10.1126/stke.2002.132.pe25. Sci STKE. 2002. PMID: 12011495 Review. - Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis.
Wuichet K, Alexander RP, Zhulin IB. Wuichet K, et al. Methods Enzymol. 2007;422:1-31. doi: 10.1016/S0076-6879(06)22001-9. Methods Enzymol. 2007. PMID: 17628132 Free PMC article. Review.
Cited by
- Novel methods for analysing bacterial tracks reveal persistence in Rhodobacter sphaeroides.
Rosser G, Fletcher AG, Wilkinson DA, de Beyer JA, Yates CA, Armitage JP, Maini PK, Baker RE. Rosser G, et al. PLoS Comput Biol. 2013 Oct;9(10):e1003276. doi: 10.1371/journal.pcbi.1003276. Epub 2013 Oct 24. PLoS Comput Biol. 2013. PMID: 24204227 Free PMC article. - Swarming bacteria migrate by Lévy Walk.
Ariel G, Rabani A, Benisty S, Partridge JD, Harshey RM, Be'er A. Ariel G, et al. Nat Commun. 2015 Sep 25;6:8396. doi: 10.1038/ncomms9396. Nat Commun. 2015. PMID: 26403719 Free PMC article. - Methods and Measures for Investigating Microscale Motility.
Bondoc-Naumovitz KG, Laeverenz-Schlogelhofer H, Poon RN, Boggon AK, Bentley SA, Cortese D, Wan KY. Bondoc-Naumovitz KG, et al. Integr Comp Biol. 2023 Dec 29;63(6):1485-1508. doi: 10.1093/icb/icad075. Integr Comp Biol. 2023. PMID: 37336589 Free PMC article. Review. - Bacterial tethering analysis reveals a "run-reverse-turn" mechanism for Pseudomonas species motility.
Qian C, Wong CC, Swarup S, Chiam KH. Qian C, et al. Appl Environ Microbiol. 2013 Aug;79(15):4734-43. doi: 10.1128/AEM.01027-13. Epub 2013 May 31. Appl Environ Microbiol. 2013. PMID: 23728820 Free PMC article. - Tumor spheroids under perfusion within a 3D microfluidic platform reveal critical roles of cell-cell adhesion in tumor invasion.
Huang YL, Ma Y, Wu C, Shiau C, Segall JE, Wu M. Huang YL, et al. Sci Rep. 2020 Jun 15;10(1):9648. doi: 10.1038/s41598-020-66528-2. Sci Rep. 2020. PMID: 32541776 Free PMC article.
References
- Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. - PubMed
- Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. - PubMed
- Ben Jacob, E., I. Becker, Y. Shapira, and H. Levine. 2004. Bacterial linguistic communication and social intelligence. Trends Microbiol. 12:366-372. - PubMed
- Berg, H. C. 2004. E. coli in motion. Springer-Verlag, Berlin, Germany.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials