MEME: discovering and analyzing DNA and protein sequence motifs - PubMed (original) (raw)
MEME: discovering and analyzing DNA and protein sequence motifs
Timothy L Bailey et al. Nucleic Acids Res. 2006.
Abstract
MEME (Multiple EM for Motif Elicitation) is one of the most widely used tools for searching for novel 'signals' in sets of biological sequences. Applications include the discovery of new transcription factor binding sites and protein domains. MEME works by searching for repeated, ungapped sequence patterns that occur in the DNA or protein sequences provided by the user. Users can perform MEME searches via the web server hosted by the National Biomedical Computation Resource (http://meme.nbcr.net) and several mirror sites. Through the same web server, users can also access the Motif Alignment and Search Tool to search sequence databases for matches to motifs encoded in several popular formats. By clicking on buttons in the MEME output, users can compare the motifs discovered in their input sequences with databases of known motifs, search sequence databases for matches to the motifs and display the motifs in various formats. This article describes the freely accessible web server and its architecture, and discusses ways to use MEME effectively to find new sequence patterns in biological sequences and analyze their significance.
Figures
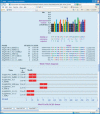
Figure 1
Sample MEME output.This portion of an MEME HTML output form shows a protein motif that MEME has discovered in the input sequences. The sites identified as belonging to the motif are indicated, and above them is the ‘consensus’ of the motif and a color-coded bar graph showing the conservation of each position in the motif. Some of the hyperlinked buttons that allow the motif to be viewed and analyzed in other ways can be seen at the bottom of the screen shot.
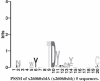
Figure 2
LOGO of protein motif. LOGOS are a visualization tool for motifs. The height of a letter indicates its relative frequency at the given position (_x_-axis) in the motif.
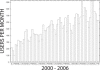
Figure 3
Usage of MEME at the NBCR web server. The plot shows the number of different users submitting jobs to the NBCR MEME web server each month since December 2000. Usage figures for March 2006 include up to March 20 only.
Similar articles
- MEME SUITE: tools for motif discovery and searching.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. Bailey TL, et al. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W202-8. doi: 10.1093/nar/gkp335. Epub 2009 May 20. Nucleic Acids Res. 2009. PMID: 19458158 Free PMC article. - STAMP: a web tool for exploring DNA-binding motif similarities.
Mahony S, Benos PV. Mahony S, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W253-8. doi: 10.1093/nar/gkm272. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478497 Free PMC article. - Discovering novel sequence motifs with MEME.
Bailey TL. Bailey TL. Curr Protoc Bioinformatics. 2002 Nov;Chapter 2:Unit 2.4. doi: 10.1002/0471250953.bi0204s00. Curr Protoc Bioinformatics. 2002. PMID: 18792935 - Discovering sequence motifs.
Bailey TL. Bailey TL. Methods Mol Biol. 2008;452:231-51. doi: 10.1007/978-1-60327-159-2_12. Methods Mol Biol. 2008. PMID: 18566768 Review. - Amino acid motifs for the identification of novel protein interactants.
Wong A, Bi C, Chi W, Hu N, Gehring C. Wong A, et al. Comput Struct Biotechnol J. 2022 Dec 10;21:326-334. doi: 10.1016/j.csbj.2022.12.012. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36582434 Free PMC article. Review.
Cited by
- Genome-wide analysis and expression profile of the bZIP gene family in Neopyropia yezoensis.
Zhu X, Gao T, Bian K, Meng C, Tang X, Mao Y. Zhu X, et al. Front Plant Sci. 2024 Oct 21;15:1461922. doi: 10.3389/fpls.2024.1461922. eCollection 2024. Front Plant Sci. 2024. PMID: 39498397 Free PMC article. - Genome-Wide Identification of the bHLH Gene Family in Rhododendron delavayi and Its Expression Analysis in Different Floral Tissues.
Dong J, Wu YW, Dong Y, Pu R, Li XJ, Lyu YM, Bai T, Zhang JL. Dong J, et al. Genes (Basel). 2024 Sep 26;15(10):1256. doi: 10.3390/genes15101256. Genes (Basel). 2024. PMID: 39457380 Free PMC article. - Predicting the role of β-GAL genes in bean under abiotic stress and genome-wide characterization of β-GAL gene family members.
Buttanri A, Kasapoğlu AG, Öner BM, Aygören AS, Muslu S, İlhan E, Yildirim E, Aydin M. Buttanri A, et al. Protoplasma. 2024 Oct 23. doi: 10.1007/s00709-024-01998-z. Online ahead of print. Protoplasma. 2024. PMID: 39441340 - Genome-wide exploration of the CONSTANS-like (COL) gene family and its potential role in regulating plant flowering time in foxtail millet (Setaria italica).
Jiang L, Li G, Shao C, Gao K, Ma N, Rao J, Miao X. Jiang L, et al. Sci Rep. 2024 Oct 18;14(1):24518. doi: 10.1038/s41598-024-74724-7. Sci Rep. 2024. PMID: 39424865 Free PMC article. - Genome-Wide Identification, Phylogenetic, and Expression Analysis of Jasmonate ZIM-Domain Gene Family in Medicago Sativa L.
Cui J, Jiang X, Li Y, Zhang L, Zhang Y, Wang X, He F, Li M, Zhang T, Kang J. Cui J, et al. Int J Mol Sci. 2024 Oct 1;25(19):10589. doi: 10.3390/ijms251910589. Int J Mol Sci. 2024. PMID: 39408917 Free PMC article.
References
- Bailey T.L., Elkan C. Unsupervised Learning of Multiple Motifs In Biopolymers Using EM. Mach. Learn. 1995;21:51–80.
- Bailey T.L., Elkan C. The value of prior knowledge in discovering motifs with MEME. In: Rawlings C., Clark D., Altman R., Hunter L., Lengauer T., Wodak S., editors. Proceedings of the Third International Conference on Intelligent Systems for Molecular biology, July; Menlo Park, CA: AAAI Press; 1995. pp. 21–29. - PubMed
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R.B., Brutlag D.L., Karp P.D., Lathrop R.H., Searls D.B., editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, August; Menlo Park, CA: AAAI Press; 1994. pp. 28–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR008605/RR/NCRR NIH HHS/United States
- R01 RR021692/RR/NCRR NIH HHS/United States
- P41 RR08605/RR/NCRR NIH HHS/United States
- R01 RR021692-01/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources