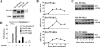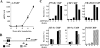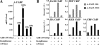Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery - PubMed (original) (raw)
Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery
Ronni Nielsen et al. Mol Cell Biol. 2006 Aug.
Abstract
Investigations of the molecular events involved in activation of genomic target genes by peroxisome proliferator-activated receptors (PPARs) have been hampered by the inability to establish a clean on/off state of the receptor in living cells. Here we show that the combination of adenoviral delivery and chromatin immunoprecipitation (ChIP) is ideal for dissecting these mechanisms. Adenoviral delivery of PPARs leads to a rapid and synchronous expression of the PPAR subtypes, establishment of transcriptional active complexes at genomic loci, and immediate activation of even silent target genes. We demonstrate that PPARgamma2 possesses considerable ligand-dependent as well as independent transactivation potential and that agonists increase the occupancy of PPARgamma2/retinoid X receptor at PPAR response elements. Intriguingly, by direct comparison of the PPARs (alpha, gamma, and beta/delta), we show that the subtypes have very different abilities to gain access to target sites and that in general the genomic occupancy correlates with the ability to activate the corresponding target gene. In addition, the specificity and potency of activation by PPAR subtypes are highly dependent on the cell type. Thus, PPAR subtype-specific activation of genomic target genes involves an intricate interplay between the properties of the subtype- and cell-type-specific settings at the individual target loci.
Figures
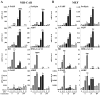
FIG. 1.
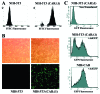
Adenoviral delivery in NIH 3T3 fibroblasts. (A) FACS analysis of CARΔ1 expression on the cell surface of NIH 3T3 and NIH 3T3 retrovirally transduced with CARΔ1. Cells were incubated with a primary antibody against CAR and a secondary fluorescein isothiocyanate-conjugated antibody, fixed with formaldehyde, and subjected to FACS analysis. (B) Recombinant AdGFP (55 PFU/cell for 8 h) were used to transduce NIH 3T3 and NIH 3T3 (CARΔ1) cells. Top frames show phase-contrast microscopy. Lower frames show GFP expression visualized by fluorescence microscopy. (C) NIH-CAR cells show homogenous uptake of AdGFP 2 h posttransduction compared to the nonselected mixed population of NIH 3T3 (CARΔ1) cells. Cells were fixed, and GFP expression was determined by FACS analysis.
FIG. 2.
Equal expression of transcriptionally active HA-PPAR subtypes in NIH-CAR cells. (A) The expression of AdHA-PPARγ2 in NIH-CAR is equal to PPARγ2 in differentiated 3T3-L1 adipocytes. Whole-cell lysates were prepared from differentiated 3T3-L1 adipocytes (day 8 and 10) and from NIH-CAR cells 8 h after transduction with AdHA-PPARγ2 (55 PFU/cell). Cell extracts were subjected to SDS-PAGE and immunoblotted using antibodies against PPARγ and TFIIB. (B) Elevated PPAR mRNA levels can be detected 2 h after adenoviral transduction. NIH-CAR cells were transduced with HA-PPARs (55 PFU/cell), and RNA was isolated at 0, 2, 4, 6, 8, and 10 h after adenoviral transduction. The PPARα, γ2, and β/δ mRNA levels were determined by real-time PCR and normalized to the corresponding TFIIB levels. (C) A distinct expression of HA-PPAR protein is present 4 h after adenoviral transduction. Whole-cell extracts were submitted to SDS-PAGE and Western blotting using antibodies against the HA epitope and TFIIB, respectively. (D) The transcriptional activity of all PPAR subtypes was verified by transient transfection of NIH-CAR cells with a multimerized PPRE luciferase reporter construct (3xPPRE-Tk-Luc) and subsequent transduction with adenoviral HA-PPARα, γ2, or β/δ (55 PFU/cell) for 12 h. Experiments were performed in the presence or absence of 30 μM WY14.643, 1 μM rosiglitazone/BRL49653, or 1 μM L-165041, respectively. Luciferase values were normalized to β-galactosidase activity and plotted relative to activity of the reporter in nontransduced NIH-CAR cells. All results are representative of a minimum of three independent experiments.
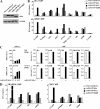
FIG. 3.
Activation of the endogenous A-FABP gene by PPARγ2. (A) PPARγ2 induces the A-FABP gene following 4 h of adenoviral transduction. NIH-CAR cells were transduced with 55 PFU/cell recombinant adenoviral PPARγ2 in the presence of 1 μM rosiglitazone/BRL49653, and RNA was purified from cells 0, 2, 4, 6, and 8 h after transduction. The A-FABP mRNA levels were determined by real-time PCR, normalized to the corresponding TFIIB levels and shown relative to the A-FABP/TFIIB level of nontransduced cells. (B) Schematic illustration of the A-FABP promoter. The relative positions of the A-FABP −5500 and −200 primers are indicated. (C) Activation of the A-FABP gene correlates with PPARγ2, CBP, TRAP220, and RNA Pol II recruitment to the A-FABP promoter and increased H3 and H4 acetylation. Chromatin was prepared 0, 2, 4, 5, and 7 h after transduction and subsequently subjected to IP using antibodies against PPARγ, CBP, TRAP220, RNA Pol II, acetylated K9 and K14 H3, and tetra-acetylated H4, respectively. Enriched DNA was analyzed using real-time PCR with primers positioned at the A-FABP PPREs (−5500) (black bars) and the proximal promoter (−200) (hatched bars). Results are shown as percent recovery relative to chromatin input. Results are representative of at least three independent experiments.
FIG. 4.
Induction of the endogenous A-FABP gene by PPARγ2 ligand-dependent and -independent transactivation. (A) PPARγ2 exhibits a pronounced ligand-independent as well as ligand-dependent transcriptional potential. RNA from NIH-CAR cells transduced for 8 h with recombinant AdHA-PPARγ2 (55 PFU/cell) in the presence of 1 μM rosiglitazone/BRL49653 and/or 1 μM GW9662 was purified. The A-FABP mRNA levels were determined by real-time PCR, normalized to the corresponding TFIIB levels and shown relative to the A-FABP/TFIIB level of nontransduced cells. (B) The ligand-dependent and -independent induction of the A-FABP gene correlates with PPARγ2/RXR, CBP, TBP, and RNA Pol II recruitment to the A-FABP promoter. Chromatin was prepared 8 h after transduction and subsequently subjected to IP with antibodies against HA tag, RXR, CBP, TBP, and RNA Pol II, respectively. DNA recovery was determined by real-time PCR with primers positioned at the A-FABP PPREs (−5500) and the proximal promoter (−200), respectively. Results are representative of at least three independent experiments.
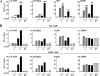
FIG. 5.
PPAR subtype-specific activation of endogenous genes in NIH 3T3 cells and MEFs. The PPARs activate endogenous target genes in a highly subtype-specific manner. NIH-CAR cells (A) and MEFs (B) were transduced with AdHA-PPARα, -γ2, or -β/δ (55 PFU/cell) in the presence of 30 μM WY14.643, 1 μM rosiglitazone/BRL49653, or 1 μM L-165041, respectively. RNA was purified 0, 4, 8, and 12 h after transduction, and expression of target genes was determined by real-time PCR, normalized to TFIIB expression and shown relative to the A-FABP/TFIIB level of nontransduced cells. A subset of genes representing different types of target genes is shown. The increase in induction (_n_-fold) of each gene at the 12-h time point is indicated. For the complete set of target genes, please refer to
http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06\_supp\_data.html
. Results are representative of at least four independent experiments.
FIG. 6.
PPAR subtype-specific binding to endogenous PPREs in the presence of specific agonists. (A) HA-PPARs are expressed at equal levels. Whole-cell extracts from NIH-CAR cells transduced with 55 PFU/cell of AdHA-PPARα, -γ2, or -β/δ for 8 h show equal protein expression of the PPAR subtypes. Proteins were separated by SDS-PAGE and immunoblotted using antibodies against the HA tag and TFIIB, respectively. (B) The ability of the different PPAR subtypes to bind to target sites in the presence of their specific agonists (Fig. 5) was determined by ChIP. Chromatin was harvested 8 h following transduction, and ChIP was performed using antibodies against the HA epitope and RXR. Relative occupancy (recovery in the presence of PPAR expression over recovery in the nontransduced cells) was determined using primers positioned at the PPREs of the indicated genes (Table 2). (C) PPARγ2 fails to further activate the transcription of the ME gene and only slightly activates that of the ACOx1 gene although receptors and cofactors are significantly recruited to the corresponding PPREs. Expression of the ME and ACOx1 genes following adenoviral transduction was determined as described in the legend of Fig. 5A. Chromatin was harvested 8 h following transduction, and ChIP was performed using antibodies against the HA tag, RXR, CBP, TRAP220, and RNA Pol II. Recovery at the ME and ACOx1 PPREs is indicated. (D) The PPAR-induced recruitment of the cofactors TRAP220 and CBP to PPREs is correlated with binding of the respective subtype to the target sites. ChIP experiments were performed as described for panel B except that antibodies against TRAP220 and CBP were used. Relative occupancy is indicated. Results are representative of three independent experiments.
FIG. 7.
The role of agonists in PPAR subtype-specific activation of endogenous target genes. (A) Activation of PPAR target genes in the presence and absence of agonists. NIH-CAR cells were transduced and treated as in described in the legend of Fig. 5A, except that the experiment was carried out in the presence as well as the absence of agonists. RNA was harvested 12 h after transduction, and expression of target genes was determined by real-time PCR, normalized to TFIIB and shown relative to the A-FABP/TFIIB level of nontransduced cells. Results are representative of at least four independent experiments. (B) Relative occupancy of HA-PPARs and RXR on PPAR target promoters in the presence and absence of agonists. NIH-CAR cells were transduced and treated as described in the legend of Fig. 6B except that the experiment was carried out in the presence as well as the absence of agonists. Relative occupancy at the indicated target genes is indicated. Results are representative of at least three independent experiments.
FIG. 8.
Cell-type-specific modulation of PPAR subtype specificity. (A) Grouping of target genes according to their relative subtype specificity in NIH-CAR fibroblasts, AML-12 hepatocytes, and MIN6 pancreatic β cells. Cells were transduced and treated with agonists, and the expression of target genes was determined as described in the legend of Fig. 5A. For a full account of target gene expression, please refer to
http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06\_supp\_data.html
. The investigated target genes are considered to be induced by a particular PPAR subtype, when the gene is activated more than threefold over background (nontransduced cells), and when the induction is at least 15% of the induction obtained by the most potent PPAR subtype. (B) Expression of selected target genes in AML-12 and MIN6 cells following transduction with HA-PPARs. Cells were transduced and treated with agonists, and expression of target genes was determined as described in the legend of Fig. 5A. (C) Relative occupancy of HA-PPARs and RXR on PPAR target promoters in AML-12 and MIN 6 cells. Cells were transduced and treated as described in the legend of Fig. 6B. Relative occupancy at the indicated target genes is indicated. Results are representative of three independent experiments.
Similar articles
- Phytoceramide and sphingoid bases derived from brewer's yeast Saccharomyces pastorianus activate peroxisome proliferator-activated receptors.
Murakami I, Wakasa Y, Yamashita S, Kurihara T, Zama K, Kobayashi N, Mizutani Y, Mitsutake S, Shigyo T, Igarashi Y. Murakami I, et al. Lipids Health Dis. 2011 Aug 24;10:150. doi: 10.1186/1476-511X-10-150. Lipids Health Dis. 2011. PMID: 21861924 Free PMC article. - The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment.
Bugge A, Grøntved L, Aagaard MM, Borup R, Mandrup S. Bugge A, et al. Mol Endocrinol. 2009 Jun;23(6):794-808. doi: 10.1210/me.2008-0236. Epub 2009 Mar 12. Mol Endocrinol. 2009. PMID: 19282365 Free PMC article. - Functional analysis of peroxisome-proliferator-responsive element motifs in genes of fatty acid-binding proteins.
Schachtrup C, Emmler T, Bleck B, Sandqvist A, Spener F. Schachtrup C, et al. Biochem J. 2004 Aug 15;382(Pt 1):239-45. doi: 10.1042/BJ20031340. Biochem J. 2004. PMID: 15130092 Free PMC article. - An overview on biological mechanisms of PPARs.
Kota BP, Huang TH, Roufogalis BD. Kota BP, et al. Pharmacol Res. 2005 Feb;51(2):85-94. doi: 10.1016/j.phrs.2004.07.012. Pharmacol Res. 2005. PMID: 15629253 Review. - Cannabinoids go nuclear: evidence for activation of peroxisome proliferator-activated receptors.
O'Sullivan SE. O'Sullivan SE. Br J Pharmacol. 2007 Nov;152(5):576-82. doi: 10.1038/sj.bjp.0707423. Epub 2007 Aug 20. Br J Pharmacol. 2007. PMID: 17704824 Free PMC article. Review.
Cited by
- Acute genome-wide effects of rosiglitazone on PPARγ transcriptional networks in adipocytes.
Haakonsson AK, Stahl Madsen M, Nielsen R, Sandelin A, Mandrup S. Haakonsson AK, et al. Mol Endocrinol. 2013 Sep;27(9):1536-49. doi: 10.1210/me.2013-1080. Epub 2013 Jul 24. Mol Endocrinol. 2013. PMID: 23885096 Free PMC article. - Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading.
Madsen MS, Siersbæk R, Boergesen M, Nielsen R, Mandrup S. Madsen MS, et al. Mol Cell Biol. 2014 Mar;34(6):939-54. doi: 10.1128/MCB.01344-13. Epub 2013 Dec 30. Mol Cell Biol. 2014. PMID: 24379442 Free PMC article. - CIDEC/FSP27 is regulated by peroxisome proliferator-activated receptor alpha and plays a critical role in fasting- and diet-induced hepatosteatosis.
Langhi C, Baldán Á. Langhi C, et al. Hepatology. 2015 Apr;61(4):1227-38. doi: 10.1002/hep.27607. Epub 2015 Mar 9. Hepatology. 2015. PMID: 25418138 Free PMC article. - The KDM5 family is required for activation of pro-proliferative cell cycle genes during adipocyte differentiation.
Brier AB, Loft A, Madsen JGS, Rosengren T, Nielsen R, Schmidt SF, Liu Z, Yan Q, Gronemeyer H, Mandrup S. Brier AB, et al. Nucleic Acids Res. 2017 Feb 28;45(4):1743-1759. doi: 10.1093/nar/gkw1156. Nucleic Acids Res. 2017. PMID: 27899593 Free PMC article. - PPARγ and NF-κB regulate the gene promoter activity of their shared repressor, TNIP1.
Gurevich I, Zhang C, Encarnacao PC, Struzynski CP, Livings SE, Aneskievich BJ. Gurevich I, et al. Biochim Biophys Acta. 2012 Jan;1819(1):1-15. doi: 10.1016/j.bbagrm.2011.09.006. Epub 2011 Oct 7. Biochim Biophys Acta. 2012. PMID: 22001530 Free PMC article.
References
- Albrektsen, T., K. S. Frederiksen, W. E. Holmes, E. Boel, K. Taylor, and J. Fleckner. 2002. Novel genes regulated by the insulin sensitizer rosiglitazone during adipocyte differentiation. Diabetes 51:1042-1051. - PubMed
- Aoyama, T., J. M. Peters, N. Iritani, T. Nakajima, K. Furihata, T. Hashimoto, and F. J. Gonzalez. 1998. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J. Biol. Chem. 273:5678-5684. - PubMed
- Bildirici, I., C. R. Roh, W. T. Schaiff, B. M. Lewkowski, D. M. Nelson, and Y. Sadovsky. 2003. The lipid droplet-associated protein adipophilin is expressed in human trophoblasts and is regulated by peroxisomal proliferator-activated receptor-gamma/retinoid X receptor. J. Clin. Endocrinol. Metab. 88:6056-6062. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources