Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation - PubMed (original) (raw)
Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation
Fernanda Agius et al. Proc Natl Acad Sci U S A. 2006.
Abstract
DNA methylation is a stable epigenetic mark for transcriptional gene silencing in diverse organisms including plants and many animals. In contrast to the well characterized mechanism of DNA methylation by methyltransferases, the mechanisms and function of active DNA demethylation have been controversial. Genetic evidence suggested that the DNA glycosylase domain-containing protein ROS1 of Arabidopsis is a putative DNA demethylase, because loss-of-function ros1 mutations cause DNA hypermethylation and enhance transcriptional gene silencing. We report here the biochemical characterization of ROS1 and the effect of its overexpression on the DNA methylation of target genes. Our data suggest that the DNA glycosylase activity of ROS1 removes 5-methylcytosine from the DNA backbone and then its lyase activity cleaves the DNA backbone at the site of 5-methylcytosine removal by successive beta- and delta-elimination reactions. Overexpression of ROS1 in transgenic plants led to a reduced level of cytosine methylation and increased expression of a target gene. These results demonstrate that ROS1 is a 5-methylcytosine DNA glycosylase/lyase important for active DNA demethylation in Arabidopsis.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
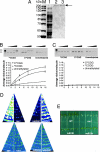
Wild-type ROS1 (wtROS1) but not mutated ROS1 (mROS1) is functional in plants and have nicking activity on methylated plasmid DNA. (A) Purification of recombinant MBP-ROS1 and MBP-mROS1 fusion proteins. Shown is an SDS/PAGE gel stained with Coomassie blue. M, molecular mass markers; lane 1, Lysate of E. coli cells after induction of MBP-ROS1; lane 2, purified MBP-ROS1; and lane 3, purified MBP-mROS1. (B and C) DNA nicking activity. Purified closed circular (CC) plasmid DNA was incubated with increasing amounts of ROS1 (B) and mROS1 (C), and the reaction mixture resolved on an agarose gel. Control reaction with unmethylated plasmids was carried out in parallel. The plots represent quantification of DNA nicking activity. The average number of nicks per plasmid was estimated from the fraction of open circular form (OC). (D) Complementation of ros1 mutant by ectopic expression of wtROS1 and mROS1. Seedlings grown on MS medium for 10 days were treated with at 4°C for 48 h before the luminescence images were taken. (E) Kanamycin sensitivity of ros1 seedlings transformed with wtROS1 or mROS1. Seeds of wild type, ros1, and ros1 transformed with wtROS1 and mROS1 were germinated on MS medium supplemented with kanamycin (35 μg/ml).
Fig. 2.
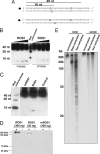
5-methylcytosine DNA glycosylase activity of ROS1 on oligonucleotide and promoter DNA substrates. (A) Sequences of methylated double stranded DNA oligonucleotides. The dot denotes digoxigenin labeling. Methylated cytosine is underlined. (B) Methyl-DNA cleavage activity of ROS1 using the substrates shown in A. (C) Comparison of wild-type ROS1 and mROS1 activity using the second substrate (5mCCGG) shown in A. Control, no enzyme. (D) NaBH4 trapping of the Schiff base reaction intermediate. Double-stranded oligonucleotide containing 5mCCGG was incubated with purified ROS1 or mROS1 in the absence (−) or presence (+) of NaBH4. Cross-linked enzyme-susbtrate complex was visualized by chemiluminescence detection after SDS/PAGE and transfer to nylon membrane. (E) A 600-bp fragment of the RD29A promoter methylated with either MspI or SssI methylase was used as a substrate in the demethylation reaction. The reaction product was resolved on a denaturing polyacrylamide gel, transferred to a nylon membrane, and hybridized with a labeled RD29A promoter cDNA probe. An unmethylated fragment of RD29A promoter was used as negative control.
Fig. 3.
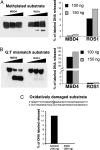
ROS1 has low activity against mismatched DNA and no activity against damaged DNA. Radiolabeled double-stranded oligonucleotide containing 5mCCGG (A) or G/T mismatch (B) were incubated with purified ROS1 or MBD4 recombinant protein. The reaction products were separated on 17% denaturing polyacrylamide gels. The released product as a percentage of total labeled DNA is shown in the graph on the right. (C) Radiolabeled double-stranded oligonucleotide containing 8-oxoG was incubated with ROS1 or AtOGG1 recombinant protein as described in Materials and Methods. The graph shows the released product as a percentage of total labeled DNA.
Fig. 4.
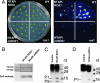
5-methylcytosine DNA glycosylase activity of ROS1 purified from plants. (A) Complementation of ros1 mutant by ectopic expression of NTAPi-ROS1 and NTAPi-mROS1. Seedlings were treated with cold (4°C for 48 h) then the luminescence image was taken. (B) SDS/PAGE and Western blot analysis of NTAPi-ROS1 and NTAPi-mROS1 proteins using antibodies against the TAP tag. (C and D) Double-stranded oligonucleotide containing 5mCCGG was incubated with NTAPi-ROS1 (C) or NTAPi-mROS1 (D). The reactions were carried out with NTAPi-ROS1/mROS1 protein immobilized on IgG-Sepharose beads. The putative β and βδ elimination products are indicated by P1 and P2, respectively.
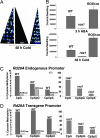
Fig. 5.
Luminescence and DNA methylation phenotype of ROS1 overexpression plants. (A and B) Luminescence was analyzed after 100 μM ABA treatment for 3 h or cold treatment (4°C) for 48 h. (C and D) DNA methylation analysis of the endogenous (C) and transgene (D) RD29A promoter by bisulfite sequencing. Twenty clones were sequenced for each genotype.
Similar articles
- Regulatory link between DNA methylation and active demethylation in Arabidopsis.
Lei M, Zhang H, Julian R, Tang K, Xie S, Zhu JK. Lei M, et al. Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3553-7. doi: 10.1073/pnas.1502279112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733903 Free PMC article. - The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis.
Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. Zhu J, et al. Curr Biol. 2007 Jan 9;17(1):54-9. doi: 10.1016/j.cub.2006.10.059. Curr Biol. 2007. PMID: 17208187 - Preventing transcriptional gene silencing by active DNA demethylation.
Kapoor A, Agius F, Zhu JK. Kapoor A, et al. FEBS Lett. 2005 Oct 31;579(26):5889-98. doi: 10.1016/j.febslet.2005.08.039. Epub 2005 Aug 31. FEBS Lett. 2005. PMID: 16162337 Review. - DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases.
Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T. Morales-Ruiz T, et al. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6853-8. doi: 10.1073/pnas.0601109103. Epub 2006 Apr 19. Proc Natl Acad Sci U S A. 2006. PMID: 16624880 Free PMC article. - Active DNA demethylation in plants: 20 years of discovery and beyond.
Zhang H, Gong Z, Zhu JK. Zhang H, et al. J Integr Plant Biol. 2022 Dec;64(12):2217-2239. doi: 10.1111/jipb.13423. J Integr Plant Biol. 2022. PMID: 36478523 Review.
Cited by
- An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis.
Zheng Z, Xing Y, He XJ, Li W, Hu Y, Yadav SK, Oh J, Zhu JK. Zheng Z, et al. Plant J. 2010 Apr 1;62(1):92-9. doi: 10.1111/j.1365-313X.2010.04130.x. Epub 2010 Jan 6. Plant J. 2010. PMID: 20059743 Free PMC article. - ROS1-Dependent DNA Demethylation Is Required for ABA-Inducible NIC3 Expression.
Kim JS, Lim JY, Shin H, Kim BG, Yoo SD, Kim WT, Huh JH. Kim JS, et al. Plant Physiol. 2019 Apr;179(4):1810-1821. doi: 10.1104/pp.18.01471. Epub 2019 Jan 28. Plant Physiol. 2019. PMID: 30692220 Free PMC article. - Somatic DNA demethylation generates tissue-specific methylation states and impacts flowering time.
Williams BP, Bechen LL, Pohlmann DA, Gehring M. Williams BP, et al. Plant Cell. 2022 Mar 29;34(4):1189-1206. doi: 10.1093/plcell/koab319. Plant Cell. 2022. PMID: 34954804 Free PMC article. - Arabidopsis C-terminal domain phosphatase-like 1 functions in miRNA accumulation and DNA methylation.
Jeong IS, Aksoy E, Fukudome A, Akhter S, Hiraguri A, Fukuhara T, Bahk JD, Koiwa H. Jeong IS, et al. PLoS One. 2013 Sep 18;8(9):e74739. doi: 10.1371/journal.pone.0074739. eCollection 2013. PLoS One. 2013. PMID: 24058624 Free PMC article. - Regulation of beta 4-integrin expression by epigenetic modifications in the mammary gland and during the epithelial-to-mesenchymal transition.
Yang X, Pursell B, Lu S, Chang TK, Mercurio AM. Yang X, et al. J Cell Sci. 2009 Jul 15;122(Pt 14):2473-80. doi: 10.1242/jcs.049148. Epub 2009 Jun 23. J Cell Sci. 2009. PMID: 19549682 Free PMC article.
References
- Bird A. P., Wolffe A. P. Cell. 1999;99:451–454. - PubMed
- Rangwala S. H., Richards E. Curr. Opin. Genet. Dev. 2004;14:686–691. - PubMed
- Chan S. W., Henderson I. R., Jacobsen S. E. Nat. Rev. Genet. 2005;6:351–360. - PubMed
- Robertson K. D. Nat. Rev. Genet. 2005;6:597–610. - PubMed
- Feinberg A. P., Ohlsson R., Henikoff S. Nat. Rev. Genet. 2006;7:21–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources