Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts - PubMed (original) (raw)
Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts
Fulvio Reggiori et al. Autophagy. 2005 Jul.
Abstract
Autophagy is a degradative process conserved among eukaryotic cells. It allows the elimination of cytoplasm including aberrant protein aggregates and damaged organelles. Accordingly, it is implicated in normal developmental processes and also serves a protective role in tumor suppression and elimination of invading pathogens, whereas defects in autophagy are associated with various human diseases including cancer and neurodegeneration. Atg proteins mediate the sequestration event that occurs at the preautophagosomal structure (PAS) by catalyzing the formation of double-membrane vesicles, termed autophagosomes. In Saccharomyces cerevisiae, the integral membrane protein Atg9 that is required for autophagy cycles through the PAS. Here, we demonstrate that Atg9 shuttles between this location and mitochondria. These data support a new model where mitochondria may provide at least part of the autophagosomal lipids and suggest a novel cellular function for this well-studied organelle.
Figures
Figure 1
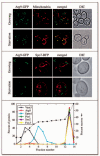
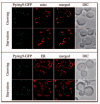
Atg9 localizes to mitochondria. (A) Atg9 distributes to mitochondria. The Atg9-GFP (FRY162) strain was grown to OD600 = 1.0 in YPD or starved in SD-N for 1 h. MitoFluor Red was added prior to imaging. DIC, differential interference contrast. (B) Atg9 does not localize to the ER. The Atg9-GFP strain carrying the SPO7RFP plasmid was grown and imaged as in (A). (C) A population of Atg9 fractionates with mitochondria. A P13 pellet fraction was separated on a sucrose density gradient as described in Materials and Methods. In total, 14 fractions were collected from the top of the gradient, resolved by SDS-PAGE, and membranes were probed with antisera to porin, Vam3, Sso2, Dpm1, Fox3 and Atg9.
Figure 2
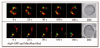
Clusters of Atg9 undergo dynamic movement on mitochondria. The Atg9-GFP (FRY162) strain was grown to an early log phase and stained with MitoFluor Red. Images of the same cells were collected every 5 s for 2.5 min. (A) The movement of three different Atg9-GFP-containing punctate structures within the mitochondrial boundaries is illustrated, and (B) a putative budding or fission event of an Atg9 cluster from mitochondria is shown. The white arrows mark a constant point for reference. For the complete movies see the supplemental data (Supplemental Video S2 and S3).
Figure 3
Atg9 cycles between mitochondria and the PAS. The ATG9-YFP _atg1_Δ (FRY138) strain was cotransformed with the pTS470 (CFP-Ape1) and pATG1ts415 plasmids. Transformants were grown at 24°C to an early log phase, incubated in the presence of MitoFluor Red for 40 min and photographed. The same cells were then transferred to 35°C for 1 h and imaged again.
Figure 4
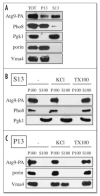
Atg9 is associated with membranous structures. (A) Most of the Atg9 is in a low speed supernatant fraction. Spheroplasts from Atg9-PA _pep4_Δ (FRY172) cells were lysed osmotically and fractionated at 13,000 xg for 12 min into total (TOT), supernatant (S13), and pellet (P13) fractions. After trichloroacetic acid precipitation, proteins were separated by SDS-PAGE and revealed by Western-blot using antibodies and antisera against protein A (PA), Pgk1, porin, Pho8 and Vma4. (B) Atg9 is always associated with membranes. S13 and P13 fractions prepared as in (A) were subjected to the following treatments: lysis buffer (-), 1 M KCl (KCl) and 1% Triton X-100 (TX-100) before being centrifuged at 100,000 xg for 30 min. Proteins from the supernatant (S100) and pellet (P100) fractions were analyzed as in (A).
Figure 5
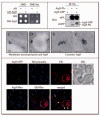
Atg9 is in a complex with itself. (A) A yeast two-hybrid analysis reveals that Atg9 can bind itself. pGAD-ATG9, pGDBU-ATG9 and the vectors without inserts were cotransformed in the appropriate combinations into the PJ69-4A strain. Interactions were monitored by the ability of cells to grow on plates lacking histidine (-his). (B) Atg9 affinity isolation. The Atg9-PA _pep4_Δ (FRY172) strain carrying either an empty plasmid (pRS416) or one expressing Atg9-GFP (pCuGFPATG9(416)), and the _pep4_Δ (TVY1) strain expressing Atg9-GFP were used to prepare detergent-solubilized extracts. Protein A fusions together with the associated proteins were affinity purified (IP) with IgG sepharose beads. Eluted polypeptides were separated by SDS-PAGE and visualized by immunoblotting with antibodies to PA (Dako, Glostrup, Denmark) and GFP (Covance Research Products, Berkeley, CA). (C-F) The Atg9-3HA (PSY278) strain expressing the _ATG9_-triple HA fusion under the control of the authentic ATG9 promoter was grown at 30°C in YPD medium to mid log phase and cells prepared for immuno-electron microscopy as described in Materials and Methods. (C) and (D) represent images of Atg9 (6 nm gold particles) colocalizing with porin (12 nm gold particles) around the mitochondrial surface, whereas (E) and (F) illustrate examples of cytosolic Atg9 clusters. White bars, 0.1 μm. (G) Atg9 is not concentrated to the MAMs. The ATG9-GFP (FRY162) strain carrying both the promSPO7RFP424 and pYES-mtBFP plasmids was grown overnight to an early log phase in a selective medium with galactose as the sole carbon source before being transferred for 3 h to medium containing glucose. Cells were then fixed as described in Materials and Methods and imaged. Mito, mitochondria. Open and closed arrowheads mark the locations of Atg9 colocalizing or not colocalizing with MAMs, respectively.
Figure 6
The P. pastoris Atg9 orthologue also localizes to mitochondria. (A) PpAtg9 is partially distributed to mitochondria. The PpATG9-GFP (FRPPY002) strain was grown to early log phase or starved in SD-N medium for 1 h, incubated in the presence of 0.05 μM MitoTracker Red CMXRos for 30 min and fixed as described in Materials and Methods before imaging. (B) PpAtg9 does not localize to the ER. The DsRedHDEL PpATG9-GFP (FRPPY003) strain was grown, fixed and imaged as in (A).
Figure 7
Putative model for Atg9 trafficking in yeast S. cerevisiae. Atg9-containing membranes are derived from mitochondria and become an isolation membrane or phagophore by acquiring Atg16 and Atg5. The covalent linkage of Atg12 to Atg5, and other events that follow autophagic induction trigger autophagosome formation. Additional cytosolic Atg12-Atg5 conjugate are recruited to this compartment and start to redistribute, becoming concentrated mostly on the external lipid bilayer. The Atg12-Atg5-Atg16 complex then forms larger oligomers that dictate the elongation of the phagophore by conveying Atg8-containing membranes and possibly additional Atg9 clusters to the crescent structure. Once the autophagosome is completed, the Atg12-Atg5 conjugate and Atg16 dissociate from this structure whereas Atg4 releases the Atg8 on the external lipid bilayer into the cytosol by proteolytically cleaving this molecule from the lipid moiety. At the same time, Atg9 is recycled back to mitochondria for reuse. These final uncoating events allow the autophagosome to fuse with the lysosome/vacuole.
Similar articles
- Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast.
He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. He C, et al. J Cell Biol. 2006 Dec 18;175(6):925-35. doi: 10.1083/jcb.200606084. J Cell Biol. 2006. PMID: 17178909 Free PMC article. - Atg27 is required for autophagy-dependent cycling of Atg9.
Yen WL, Legakis JE, Nair U, Klionsky DJ. Yen WL, et al. Mol Biol Cell. 2007 Feb;18(2):581-93. doi: 10.1091/mbc.e06-07-0612. Epub 2006 Nov 29. Mol Biol Cell. 2007. PMID: 17135291 Free PMC article. - Atg17 recruits Atg9 to organize the pre-autophagosomal structure.
Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Sekito T, et al. Genes Cells. 2009 May;14(5):525-38. doi: 10.1111/j.1365-2443.2009.01299.x. Epub 2009 Apr 13. Genes Cells. 2009. PMID: 19371383 - Atg9 trafficking in the yeast Saccharomyces cerevisiae.
Mari M, Reggiori F. Mari M, et al. Autophagy. 2007 Mar-Apr;3(2):145-8. doi: 10.4161/auto.3608. Epub 2007 Mar 21. Autophagy. 2007. PMID: 17204846 Review. - Atg9 trafficking in Mammalian cells.
Webber JL, Young AR, Tooze SA. Webber JL, et al. Autophagy. 2007 Jan-Feb;3(1):54-6. doi: 10.4161/auto.3419. Epub 2007 Jan 20. Autophagy. 2007. PMID: 17102588 Review.
Cited by
- The protective roles of autophagy in ischemic preconditioning.
Yan WJ, Dong HL, Xiong LZ. Yan WJ, et al. Acta Pharmacol Sin. 2013 May;34(5):636-43. doi: 10.1038/aps.2013.18. Epub 2013 Apr 22. Acta Pharmacol Sin. 2013. PMID: 23603984 Free PMC article. Review. - The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae.
Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. Reggiori F, et al. Mol Biol Cell. 2005 Dec;16(12):5843-56. doi: 10.1091/mbc.e05-07-0629. Epub 2005 Oct 12. Mol Biol Cell. 2005. PMID: 16221887 Free PMC article. - The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy.
Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. Yen WL, et al. J Cell Biol. 2010 Jan 11;188(1):101-14. doi: 10.1083/jcb.200904075. J Cell Biol. 2010. PMID: 20065092 Free PMC article. - Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise.
Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Wohlgemuth SE, et al. Exp Gerontol. 2010 Feb;45(2):138-48. doi: 10.1016/j.exger.2009.11.002. Epub 2009 Nov 10. Exp Gerontol. 2010. PMID: 19903516 Free PMC article. - Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast.
He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. He C, et al. J Cell Biol. 2006 Dec 18;175(6):925-35. doi: 10.1083/jcb.200606084. J Cell Biol. 2006. PMID: 17178909 Free PMC article.
References
- Levine B. Eating oneself and uninvited guests; autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. - PubMed
- Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. - PubMed
- Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases