Auxin signaling - PubMed (original) (raw)
Review
Auxin signaling
Marcel Quint et al. Curr Opin Plant Biol. 2006 Oct.
Abstract
Auxin regulates a host of plant developmental and physiological processes, including embryogenesis, vascular differentiation, organogenesis, tropic growth, and root and shoot architecture. Genetic and biochemical studies carried out over the past decade have revealed that much of this regulation involves the SCF(TIR1/AFB)-mediated proteolysis of the Aux/IAA family of transcriptional regulators. With the recent finding that the TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN SIGNALING F-BOX (AFB) proteins also function as auxin receptors, a potentially complete, and surprisingly simple, signaling pathway from perception to transcriptional response is now before us. However, understanding how this seemingly simple pathway controls the myriad of specific auxin responses remains a daunting challenge, and compelling evidence exists for SCF(TIR1/AFB)-independent auxin signaling pathways.
Figures
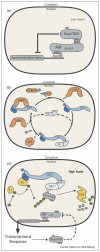
Auxin regulation of gene expression. (a) Under sub-threshold auxin concentrations the Aux/IAA proteins heterodimerize with the ARF transcription factors, thereby repressing auxin-inducible gene expression. (b) The active SCFTIR1 complex, containing RUB-modified CUL1, is shown at the bottom. The CSN complex can cleave the RUB modifier from CUL1, thus facilitating CAND1 binding to CUL1 and SCF disassembly. Conjugation of RUB to CUL1 by the AXR1-ECR1 and RCE1 enzymes might free CUL1 from CAND1, promoting re-assembly of the active complex. Genetic studies have shown that all of the components depicted in this figure are required for optimal SCFTIR1 activity in vivo. (c) Auxin binding to the TIR1/AFB receptors promotes the recruitment of Aux/IAA proteins to the SCF complex. Subsequent Aux/IAA ubiquitinylation and proteasome-mediated degradation results in a decline in Aux/IAA protein levels, thus de-repressing auxin-inducible gene expression. In addition to genes responsible for specific auxin responses, the Aux/IAA genes themselves are auxin-inducible. This might represent a negative feedback loop that ensures a transient response, with the nascent Aux/IAA proteins attenuating the signaling pathway as auxin levels fall by restoring repression of the ARF transcription factors. DBD, DNA-binding domain; E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; U, ubiquitin; R, RUB. *,AFB1, AFB2, or AFB3.
Similar articles
- Auxin receptors and plant development: a new signaling paradigm.
Mockaitis K, Estelle M. Mockaitis K, et al. Annu Rev Cell Dev Biol. 2008;24:55-80. doi: 10.1146/annurev.cellbio.23.090506.123214. Annu Rev Cell Dev Biol. 2008. PMID: 18631113 Review. - Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development.
Vain T, Raggi S, Ferro N, Barange DK, Kieffer M, Ma Q, Doyle SM, Thelander M, Pařízková B, Novák O, Ismail A, Enquist PA, Rigal A, Łangowska M, Ramans Harborough S, Zhang Y, Ljung K, Callis J, Almqvist F, Kepinski S, Estelle M, Pauwels L, Robert S. Vain T, et al. Proc Natl Acad Sci U S A. 2019 Mar 26;116(13):6463-6472. doi: 10.1073/pnas.1809037116. Epub 2019 Mar 8. Proc Natl Acad Sci U S A. 2019. PMID: 30850516 Free PMC article. - Auxin-binding protein 1 is a negative regulator of the SCF(TIR1/AFB) pathway.
Tromas A, Paque S, Stierlé V, Quettier AL, Muller P, Lechner E, Genschik P, Perrot-Rechenmann C. Tromas A, et al. Nat Commun. 2013;4:2496. doi: 10.1038/ncomms3496. Nat Commun. 2013. PMID: 24051655 - The F-box protein TIR1 is an auxin receptor.
Dharmasiri N, Dharmasiri S, Estelle M. Dharmasiri N, et al. Nature. 2005 May 26;435(7041):441-5. doi: 10.1038/nature03543. Nature. 2005. PMID: 15917797 - SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development.
Salehin M, Bagchi R, Estelle M. Salehin M, et al. Plant Cell. 2015 Jan;27(1):9-19. doi: 10.1105/tpc.114.133744. Epub 2015 Jan 20. Plant Cell. 2015. PMID: 25604443 Free PMC article. Review.
Cited by
- Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers.
Kobayashi Y, Kobayashi Y, Sugimoto M, Lakshmanan V, Iuchi S, Kobayashi M, Bais HP, Koyama H. Kobayashi Y, et al. Plant Physiol. 2013 Jun;162(2):732-40. doi: 10.1104/pp.113.218065. Epub 2013 Apr 26. Plant Physiol. 2013. PMID: 23624855 Free PMC article. - Characterizing seed dormancy in Epimedium brevicornu Maxim.: Development of novel chill models and determination of dormancy release mechanisms by transcriptomics.
Li P, Xiang Q, Wang Y, Dong X. Li P, et al. BMC Plant Biol. 2024 Aug 8;24(1):757. doi: 10.1186/s12870-024-05471-0. BMC Plant Biol. 2024. PMID: 39112934 Free PMC article. - Synthesis and regulation of auxin and abscisic acid in maize.
Yue K, Lingling L, Xie J, Coulter JA, Luo Z. Yue K, et al. Plant Signal Behav. 2021 Jul 3;16(7):1891756. doi: 10.1080/15592324.2021.1891756. Epub 2021 May 30. Plant Signal Behav. 2021. PMID: 34057034 Free PMC article. Review. - Low-dose 60Co-γ-ray irradiation promotes the growth of cucumber seedlings by inducing CsSAUR37 expression.
Li S, Lu K, Zhang L, Fan L, Lv W, Liu DJ, Feng G. Li S, et al. Plant Mol Biol. 2024 Sep 27;114(5):107. doi: 10.1007/s11103-024-01504-2. Plant Mol Biol. 2024. PMID: 39333431 - Expression and regulation of the early auxin-responsive Aux/IAA genes during strawberry fruit development.
Liu DJ, Chen JY, Lu WJ. Liu DJ, et al. Mol Biol Rep. 2011 Feb;38(2):1187-93. doi: 10.1007/s11033-010-0216-x. Epub 2010 Jun 19. Mol Biol Rep. 2011. PMID: 20563652
References
- Went FW, Thimann KV. Phytohormones. Macmillan; New York: 1937.
- An excellent detailed overview of auxin biology.
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous