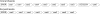A putative viral defence mechanism in archaeal cells - PubMed (original) (raw)
A putative viral defence mechanism in archaeal cells
Reidun K Lillestøl et al. Archaea. 2006 Aug.
Abstract
Clusters of regularly spaced direct repeats, separated by unconserved spacer sequences, are ubiquitous in archaeal chromosomes and occur in some plasmids. Some clusters constitute around 1% of chromosomal DNA. Similarly structured clusters, generally smaller, also occur in some bacterial chromosomes. Although early studies implicated these clusters in segregation/partition functions, recent evidence suggests that the spacer sequences derive from extrachromosomal elements, and, primarily, viruses. This has led to the proposal that the clusters provide a defence against viral propagation in cells, and that both the mode of inhibition of viral propagation and the mechanism of adding spacer-repeat units to clusters, are dependent on RNAs transcribed from the clusters. Moreover, the putative inhibitory apparatus (piRNA-based) may be evolutionarily related to the interference RNA systems (siRNA and miRNA), which are common in eukarya. Here, we analyze all the current data on archaeal repeat clusters and provide some new insights into their diverse structures, transcriptional properties and mode of structural development. The results are consistent with larger cluster transcripts being processed at the centers of the repeat sequences and being further trimmed by exonucleases to yield a dominant, intracellular RNA species, which corresponds approximately to the size of a spacer. Furthermore, analysis of the extensive clusters of Sulfolobus solfataricus strains P1 and P2B provides support for the presence of a flanking sequence adjoining a cluster being a prerequisite for the incorporation of new spacer-repeat units, which occurs between the flanking sequence and the cluster. An archaeal database summarizing the data will be maintained at http://dac.molbio.ku.dk/dbs/SRSR/.
Figures
Figure 1.
(A) A map of mthe-124 from M. thermoautotrophicus showing the locations of duplicated spacer-repeat units, or groups of units, labeled 1–5. Each triangle represents a spacer-repeat unit. Duplicated spacer-repeat units are shaded in gray. (B) Schematic representation of the six clusters which occur in the genome of_S. solfataricus_ P2B (She et al. 2001). Each triangle represents a spacer-repeat unit. The colored triangles are coded to indicate the archaeal viruses, plasmids or chromosomes (S. solfataricus or A. fulgidus) that yield good matches with the spacer sequence. (C) A corresponding scheme is presented for three of the six clusters (ssol-95, ssol-32 and ssol-7) that are present in the chromosome of the closely related strain S. solfataricus P1 (Accession numbers: DQ831675, DQ831676 and DQ831677). Open triangles denote those spacer-repeat units that differ in sequence between the strains P1 and P2A. Abbreviation: del. indicates the two sites where deletion of several spacer-repeat units has occurred in strain P1 (or where insertions have occurred in strain P2B).
Figure 2.
(A) Alignment of the five conserved flanking sequences adjoining the first repeat (in bold type) of five clusters of the three_Methanosarcina_ strains. A putative TATA-like box is outlined. (B) Alignment of two flanking regions of the_P. abyssi_ genome where paby-1 appears to exhibit a defective start site.
Figure 3.
Typical composition and orientation of the major gene components of superoperons which are closely linked to repeat clusters in crenarchaea and euryarchaea. The cas genes and their COG identities are given in Tables 4 and 5.
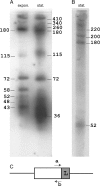
Figure 4.
Northern blotting analyses of RNA transcripts obtained from the cluster saci-4 of S. acidocaldarius. (A) Transcripts from one strand were detected in total RNA extracted from cells grown to exponential (expon.) or stationary (stat.) phase. (B) Transcripts were observed from the complementary DNA strand at stationary phase. The approximate estimated nucleotide lengths of the RNA products are given. The minimal detection limit, using 26-nt. probes, was estimated at 15–16 nucleotides. (C) Denotes the location of the primers “a” and “b,” and “L” indicates the location of the putative transcriptional leader within the flanking sequence.
Similar articles
- Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes.
Peng X, Brügger K, Shen B, Chen L, She Q, Garrett RA. Peng X, et al. J Bacteriol. 2003 Apr;185(8):2410-7. doi: 10.1128/JB.185.8.2410-2417.2003. J Bacteriol. 2003. PMID: 12670964 Free PMC article. - CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties.
Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. Lillestøl RK, et al. Mol Microbiol. 2009 Apr;72(1):259-72. doi: 10.1111/j.1365-2958.2009.06641.x. Epub 2009 Feb 23. Mol Microbiol. 2009. PMID: 19239620 - Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism.
Shah SA, Hansen NR, Garrett RA. Shah SA, et al. Biochem Soc Trans. 2009 Feb;37(Pt 1):23-8. doi: 10.1042/BST0370023. Biochem Soc Trans. 2009. PMID: 19143596 - CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea.
Sorek R, Kunin V, Hugenholtz P. Sorek R, et al. Nat Rev Microbiol. 2008 Mar;6(3):181-6. doi: 10.1038/nrmicro1793. Nat Rev Microbiol. 2008. PMID: 18157154 Review. - CRISPR/Cas, the immune system of bacteria and archaea.
Horvath P, Barrangou R. Horvath P, et al. Science. 2010 Jan 8;327(5962):167-70. doi: 10.1126/science.1179555. Science. 2010. PMID: 20056882 Review.
Cited by
- Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.
Charpentier E, Richter H, van der Oost J, White MF. Charpentier E, et al. FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review. - CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats.
Grissa I, Vergnaud G, Pourcel C. Grissa I, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W52-7. doi: 10.1093/nar/gkm360. Epub 2007 May 30. Nucleic Acids Res. 2007. PMID: 17537822 Free PMC article. - Detection of CRISPR adaptation.
Shiriaeva A, Fedorov I, Vyhovskyi D, Severinov K. Shiriaeva A, et al. Biochem Soc Trans. 2020 Feb 28;48(1):257-269. doi: 10.1042/BST20190662. Biochem Soc Trans. 2020. PMID: 32010936 Free PMC article. Review. - A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species.
Erdmann S, Chen B, Huang X, Deng L, Liu C, Shah SA, Le Moine Bauer S, Sobrino CL, Wang H, Wei Y, She Q, Garrett RA, Huang L, Lin L. Erdmann S, et al. Extremophiles. 2014 Jan;18(1):51-60. doi: 10.1007/s00792-013-0591-z. Epub 2013 Oct 27. Extremophiles. 2014. PMID: 24163004 - Analysis of the complete genome of Fervidococcus fontis confirms the distinct phylogenetic position of the order Fervidicoccales and suggests its environmental function.
Lebedinsky AV, Mardanov AV, Kublanov IV, Gumerov VM, Beletsky AV, Perevalova AA, Bidzhieva SKh, Bonch-Osmolovskaya EA, Skryabin KG, Ravin NV. Lebedinsky AV, et al. Extremophiles. 2014 Mar;18(2):295-309. doi: 10.1007/s00792-013-0616-7. Epub 2013 Dec 24. Extremophiles. 2014. PMID: 24366681
References
- Bath C., Cukalac T., Porter K., Dyall-Smith M.L. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus . Virology. 2006;350:228–239. - PubMed
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. - PubMed
- Brügger K., Redder P., She Q., Confalonieri F., Zivanovic Y., Garrett R.A. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 2002;206:131–141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources