Microbial diversity in the deep sea and the underexplored "rare biosphere" - PubMed (original) (raw)
Microbial diversity in the deep sea and the underexplored "rare biosphere"
Mitchell L Sogin et al. Proc Natl Acad Sci U S A. 2006.
Abstract
The evolution of marine microbes over billions of years predicts that the composition of microbial communities should be much greater than the published estimates of a few thousand distinct kinds of microbes per liter of seawater. By adopting a massively parallel tag sequencing strategy, we show that bacterial communities of deep water masses of the North Atlantic and diffuse flow hydrothermal vents are one to two orders of magnitude more complex than previously reported for any microbial environment. A relatively small number of different populations dominate all samples, but thousands of low-abundance populations account for most of the observed phylogenetic diversity. This "rare biosphere" is very ancient and may represent a nearly inexhaustible source of genomic innovation. Members of the rare biosphere are highly divergent from each other and, at different times in earth's history, may have had a profound impact on shaping planetary processes.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
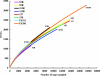
Fig. 1.
Rarefaction analysis for each sample based on best matches against the V6RefDB database. The frequency of observed best matches to V6RefDB (OTUs) for each site was used to calculate rarefaction curves with the program Analytic Rarefaction 1.3.
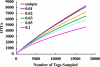
Fig. 2.
Rarefaction analysis for sample FS396 based on pairwise distance. Rarefaction is shown for OTUs that contain unique sequences and OTUs with differences that do not exceed 1%, 2%, 3%, 5%, or 10%. Rarefaction of the other seven samples showed curves with similar slopes.
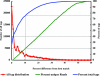
Fig. 3.
Similarity of 454 sequence tags from FS396 to the V6RefDB database. “All tag distribution” plots the number of tag sequences for all samples versus the percentage difference from the best-matching sequence in V6RefDB. “Percent unique reads” from all samples shows the percentage difference between each distinct tag sequence and its best match in V6RefDB. “Percent total tags” plots the cumulative percentage of reads in all samples at or below a given percentage difference from best matches in V6RefDB.
References
- Porter K. G., Feig Y. S. Limnol. Oceanogr. 1980;25:943–948.
- Atlas R. M., Bartha R. Microbial Ecology: Fundamentals and Applications. Redwood City, CA: Benjamin/Cummings; 1993.
- Pace N. R. Science. 1997;276:734–740. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources