Residue centrality, functionally important residues, and active site shape: analysis of enzyme and non-enzyme families - PubMed (original) (raw)
Residue centrality, functionally important residues, and active site shape: analysis of enzyme and non-enzyme families
Antonio del Sol et al. Protein Sci. 2006 Sep.
Abstract
The representation of protein structures as small-world networks facilitates the search for topological determinants, which may relate to functionally important residues. Here, we aimed to investigate the performance of residue centrality, viewed as a family fold characteristic, in identifying functionally important residues in protein families. Our study is based on 46 families, including 29 enzyme and 17 non-enzyme families. A total of 80% of these central positions corresponded to active site residues or residues in direct contact with these sites. For enzyme families, this percentage increased to 91%, while for non-enzyme families the percentage decreased substantially to 48%. A total of 70% of these central positions are located in catalytic sites in the enzyme families, 64% are in hetero-atom binding sites in those families binding hetero-atoms, and only 16% belong to protein-protein interfaces in families with protein-protein interaction data. These differences reflect the active site shape: enzyme active sites locate in surface clefts, hetero-atom binding residues are in deep cavities, while protein-protein interactions involve a more planar configuration. On the other hand, not all surface cavities or clefts are comprised of central residues. Thus, closeness centrality identifies functionally important residues in enzymes. While here we focus on binding sites, we expect to identify key residues for the integration and transmission of the information to the rest of the protein, reflecting the relationship between fold and function. Residue centrality is more conserved than the protein sequence, emphasizing the robustness of protein structures.
Figures
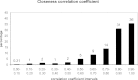
Figure 1.
Distribution over all protein families of the averaged correlation coefficients between the closeness _z_-score values for all of the aligned residues in all pairs of family members.
Figure 2.
Fragment of the 1dx4 protein family alignment. The closeness _z_-score values are shown above each residue. The _z_-score values >2.0 in at least 70% of the members of the family are highlighted in red. These are the centrally conserved residues.
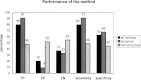
Figure 3.
Sensitivity, specificity, true positive (TP), false positive (FP), and false negative (FN) values calculated for the centrally conserved predicted residues in all families, enzyme, and non-enzyme families.
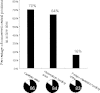
Figure 4.
Percentages of conserved central positions in all protein families located in different functional sites—catalytic, heteroatom binding, and protein–protein interaction binding. Circles below the histogram represent in each case the percentage of families with at least one conserved central residue.
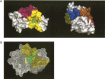
Figure 5.
(A) Representation of two views of the surface of the β-lactamase (PDB code1bsg). The deepest cavities are depicted in yellow, blue, brown, and purple. Residues from the catalytic site, located in the yellow cavity, are shown as green and red spots. (B) Mesh representation of the same protein. Residues in cyan, red, and yellow correspond to predicted centrally conserved amino acids. The residue in red is also part of the catalytic site. The remaining catalytic site residues are shown in green. Those amino acids in cyan are neighbors of the catalytic site residues. All of these residues are clustered around the same cavity (area colored in yellow).
Figure 6.
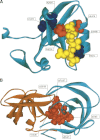
(A) Representation of the PDZ domain (PDB code 1be9). The predicted centrally conserved residues, LEU379 and PHE325, are shown in red. Both amino acids are in contact with the ligand (depicted in yellow). Residue PHE325 has been experimentally determined to be energetically coupled with residue HIS372 (key residue responsible for ligand specificity) and is part of the intramolecular signaling pathway proposed by Lockless and Ranganathan (1999). The other residues forming this pathway, ALA347 and LEU353 (colored in blue), have been suggested to participate in the allosteric communications (Lockless and Ranganathan 1999) and are in direct contact with the Cdc42 binding site of the PDZ domain in the mPar-6B protein. The functional importance of this pathway has been largely confirmed by experimental mutagenesis (Ota and Agard 2005). (B) The structure of the HIV-1 protease homodimer (PDB code1kzk). Predicted central residues are shown in red.
Similar articles
- Prediction of catalytic residues in enzymes based on known tertiary structure, stability profile, and sequence conservation.
Ota M, Kinoshita K, Nishikawa K. Ota M, et al. J Mol Biol. 2003 Apr 11;327(5):1053-64. doi: 10.1016/s0022-2836(03)00207-9. J Mol Biol. 2003. PMID: 12662930 - How accurate and statistically robust are catalytic site predictions based on closeness centrality?
Chea E, Livesay DR. Chea E, et al. BMC Bioinformatics. 2007 May 11;8:153. doi: 10.1186/1471-2105-8-153. BMC Bioinformatics. 2007. PMID: 17498304 Free PMC article. - Predicting enzyme functional surfaces and locating key residues automatically from structures.
Tseng YY, Liang J. Tseng YY, et al. Ann Biomed Eng. 2007 Jun;35(6):1037-42. doi: 10.1007/s10439-006-9241-2. Epub 2007 Feb 9. Ann Biomed Eng. 2007. PMID: 17294116 - One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions.
Nagano N, Orengo CA, Thornton JM. Nagano N, et al. J Mol Biol. 2002 Aug 30;321(5):741-65. doi: 10.1016/s0022-2836(02)00649-6. J Mol Biol. 2002. PMID: 12206759 Review. - A global analysis of function and conservation of catalytic residues in enzymes.
Ribeiro AJM, Tyzack JD, Borkakoti N, Holliday GL, Thornton JM. Ribeiro AJM, et al. J Biol Chem. 2020 Jan 10;295(2):314-324. doi: 10.1074/jbc.REV119.006289. Epub 2019 Dec 3. J Biol Chem. 2020. PMID: 31796628 Free PMC article. Review.
Cited by
- Shape and evolution of thermostable protein structure.
Coleman RG, Sharp KA. Coleman RG, et al. Proteins. 2010 Feb 1;78(2):420-33. doi: 10.1002/prot.22558. Proteins. 2010. PMID: 19731381 Free PMC article. - Exploring allosteric communication in multiple states of the bacterial ribosome using residue network analysis.
Kürkçüoğlu Ö. Kürkçüoğlu Ö. Turk J Biol. 2018 Oct 25;42(5):392-404. doi: 10.3906/biy-1802-77. eCollection 2018. Turk J Biol. 2018. PMID: 30930623 Free PMC article. - Homology Modeling and Virtual Screening Studies of Antigen MLAA-42 Protein: Identification of Novel Drug Candidates against Leukemia-An In Silico Approach.
Shehadi IA, Rashdan HRM, Abdelmonsef AH. Shehadi IA, et al. Comput Math Methods Med. 2020 Mar 16;2020:8196147. doi: 10.1155/2020/8196147. eCollection 2020. Comput Math Methods Med. 2020. PMID: 32256683 Free PMC article. - Full-scale network analysis reveals properties of the FV protein structure organization.
Ferreira-Martins AJ, Castaldoni R, Alencar BM, Ferreira MV, Nogueira T, Rios RA, Lopes TJS. Ferreira-Martins AJ, et al. Sci Rep. 2023 Jun 12;13(1):9546. doi: 10.1038/s41598-023-36528-z. Sci Rep. 2023. PMID: 37308572 Free PMC article. - Surveying the Side-Chain Network Approach to Protein Structure and Dynamics: The SARS-CoV-2 Spike Protein as an Illustrative Case.
Halder A, Anto A, Subramanyan V, Bhattacharyya M, Vishveshwara S, Vishveshwara S. Halder A, et al. Front Mol Biosci. 2020 Dec 18;7:596945. doi: 10.3389/fmolb.2020.596945. eCollection 2020. Front Mol Biosci. 2020. PMID: 33392257 Free PMC article. Review.
References
- Aloy P., Querol E., Aviles F.X., Sternberg M.J. 2001. Automated structure-based prediction of functional sites in proteins: Applications to assessing the validity of inheriting protein function from homology in genome annotation and to protein docking. J. Mol. Biol. 311 395–408. - PubMed
- Amitai G., Shemesh A., Sitbon E., Shklar M., Netanely D., Venger I., Pietrokovski S. 2004. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 344 1135–1146. - PubMed
- Atilgan A.R., Akan P., Baysal C., Vendruscolo M., Dokholyan N.V., Paci E., Karplus M. 2004. Small-world communication of residues and significance for protein dynamics. Small-world view of the amino acids that play a key role in protein folding. Biophys. J. 86 85–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources