Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis - PubMed (original) (raw)
Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis
Jian Shen et al. Appl Environ Microbiol. 2006 Aug.
Abstract
A group-specific PCR-based denaturing gradient gel electrophoresis (DGGE) method was developed and combined with group-specific clone library analysis to investigate the diversity of the Clostridium leptum subgroup in human feces. PCR products (length, 239 bp) were amplified using C. leptum cluster-specific primers and were well separated by DGGE. The DGGE patterns of fecal amplicons from 11 human individuals revealed host-specific profiles; the patterns for fecal samples collected from a child for 3 years demonstrated the structural succession of the population in the first 2 years and its stability in the third year. A clone library was constructed with 100 clones consisting of 1,143-bp inserts of 16S rRNA gene fragments that were amplified from one adult fecal DNA with one forward universal bacterial primer and one reverse group-specific primer. Eighty-six of the clones produced the 239-bp C. leptum cluster-specific amplicons, and the remaining 14 clones did not produce these amplicons but still phylogenetically belong to the subgroup. Sixty-four percent of the clones were related to Faecalibacterium prausnitzii (similarity, 97 to 99%), 6% were related to Subdoligranulum variabile (similarity, approximately 99%), 2% were related to butyrate-producing bacterium A2-207 (similarity, 99%), and 28% were not identified at the species level. The identities of most bands in the DGGE profiles for the same adult were determined by comigration analysis with the 86 clones that harbored the 239-bp group-specific fragments. Our results suggest that DGGE combined with clone library analysis is an effective technique for monitoring and analyzing the composition of this important population in the human gut flora.
Figures
FIG. 1.
(a) DGGE profiles of the fecal C. leptum group for 11 healthy individuals. Bands produced by most volunteers (at least 8 of 11 volunteers) are indicated by arrows, and the numbers correspond to band numbers in the DGGE profile of individual F shown in Fig. 3. (b) Dendrogram of the DGGE profiles shown in panel a.
FIG. 2.
Monitoring of the fecal C. leptum subgroup of a child (individual G) over time. (a) DGGE profiles of the C. leptum group for seven fecal samples collected from the healthy child during a 3-year period. Lanes 1 to 7 contained fecal samples taken on 3 June 2001, 15 July 2001, 5 August 2001, 23 August 2001, 20 March 2003, 14 April 2003, and 14 June 2004, respectively. (b) Dendrogram of the DGGE profiles shown in panel a.
FIG. 3.
Sequence analysis of representative clones belonging to different OTUs in the library and identification of dominant bands in the DGGE pattern of individual F by a comparison of the migration positions of clones to the DGGE profile. Representative clones, the species most closely related to the clones, the levels of similarity, and the richness in the clone library are indicated. A number sign indicates a DGGE band with no corresponding clone in the library. OL, clones in the library with no matching DGGE bands; c, unclassified Clostridiales; N, clones producing no amplicons when they were amplified with primers sg-Clept-F and sg-Clept-R3.
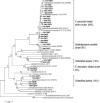
FIG. 4.
Phylogenetic tree showing the relationships between representative clones in our study and species belonging to the C. leptum cluster. The tree was constructed based on the region from base 27 to base 1181 of the 16S rRNA genes. Representative clones in the library are indicated by boldface type; clones retrieved from the GenBank database are indicated by italics, and their accession numbers are given. Bootstrap values greater than 50% are indicated at the nodes. Clostridium cellobioparum (a member of Clostridium cluster III) and Thermoanaerobacter thermocopriae and Thermoanaerobacter enthanolicus (members of Clostridium cluster V) are included as outgroups. All clones sequenced fell into the four groups indicated on the right, and the values in parentheses are the percentages of clones in the groups (a total of 100 clones). Asterisks indicate clones that do not harbor the 239-bp group-specific fragment.
References
- Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. - PubMed
- Defnoun, S., M. Labat, M. Ambrosio, J. L. Garcia, and B. K. Patel. 2000. Papillibacter cinnamivorans gen. nov., sp. nov., a cinnamate-transforming bacterium from a shea cake digester. Int. J. Syst. Evol. Microbiol. 50:1221-1228. - PubMed
- Duncan, S. H., G. L. Hold, H. J. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:2141-2146. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases