Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae - PubMed (original) (raw)
Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae
Elke Nevoigt et al. Appl Environ Microbiol. 2006 Aug.
Abstract
The strong overexpression or complete deletion of a gene gives only limited information about its control over a certain phenotype or pathway. Gene function studies based on these methods are therefore incomplete. To effect facile manipulation of gene expression across a full continuum of possible expression levels, we recently created a library of mutant promoters. Here, we provide the detailed characterization of our yeast promoter collection comprising 11 mutants of the strong constitutive Saccharomyces cerevisiae TEF1 promoter. The activities of the mutant promoters range between about 8% and 120% of the activity of the unmutated TEF1 promoter. The differences in reporter gene expression in the 11 mutants were independent of the carbon source used, and real-time PCR confirmed that these differences were due to varying levels of transcription (i.e., caused by varying promoter strengths). In addition to a CEN/ARS plasmid-based promoter collection, we also created promoter replacement cassettes. They enable genomic integration of our mutant promoter collection upstream of any given yeast gene, allowing detailed genotype-phenotype characterizations. To illustrate the utility of the method, the GPD1 promoter of S. cerevisiae was replaced by five TEF1 promoter mutants of different strengths, which allowed analysis of the impact of glycerol 3-phosphate dehydrogenase activity on the glycerol yield.
Figures
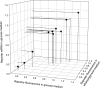
FIG. 1.
Characterization of promoter strengths in the different members of the TEF1 promoter mutant collection after retransformation of plasmids. Three different metrics are shown for each member of the promoter collection: (i) reporter expression at the protein level measured by specific fluorescence of GFP during growth in glucose as a carbon source, (ii) reporter expression at the mRNA level in cells grown in glucose medium measured by real-time PCR, and (iii) specific fluorescence of GFP during growth in ethanol-glycerol as carbon sources. All determinations were carried out using logarithmically growing cells (OD600, about 1.5). The data shown are mean values of at least two independent experiments and were normalized to the reference (unmutated TEF1 promoter; open symbol). The coefficients of variance (CV) were 7%, 19%, and 11%. These numbers are the standard deviation divided by the mean. The value for specific reporter fluorescence in ethanol-glycerol medium (19%) is highly biased by mutant 2, which has the lowest activity and a large standard deviation compared with the mean. If this point is removed, the average CV is 13.5% instead of 19%.
FIG. 2.
Multiple-sequence alignment of the unmutated TEF1 promoter (_TEF1_p; boldface) and the 11 selected TEF1 promoter mutants with graded activities. The normalized promoter strength is shown for each mutant at the beginning of each mutant sequence. The stars mark those nucleotides which remained unchanged in all promoter versions, whereas mutations are highlighted. The underlined sequences correspond to the proposed transcription factor binding sites according to the literature (3).
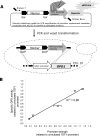
FIG. 3.
Genomic integration of five TEF1 promoter versions (the unmutated TEF1 promoter and four mutants) upstream of the GPD1 gene in S. cerevisiae. (A) Strategy of promoter replacement. (B) Activities of GPDH in the yeast transformants, depending on promoter strength. Promoter strength was calculated as the mean value of two different metrics shown in Fig. 1, i.e., relative reporter fluorescence and relative mRNA levels. Error bars indicate the range between normalized mRNA and normalized fluorescence for each promoter.
FIG. 4.
Impacts of specific GPDH activity on glycerol and biomass yields in S. cerevisiae. The values obtained by genomic integration of five TEF1 promoter versions (open symbols) and the previously known values of (i) GPD1 wild type, (i) deletion, and (iii) multicopy overexpression (closed symbols) are shown. Samples for measuring glycerol, biomass, and glucose for the calculation of yields (YX/S) were taken at the beginning of the fermentation and during logarithmic growth (optical densities were between 1.5 and 1.9). Experiments were carried out in shake flask cultures in a synthetic medium containing 2% glucose (YSC Leu−). YP/S, product yield. Error bars represent the standard deviations for two or more independent experiments.
Similar articles
- Development and characterization of a vector set with regulated promoters for systematic metabolic engineering in Saccharomyces cerevisiae.
Shen MW, Fang F, Sandmeyer S, Da Silva NA. Shen MW, et al. Yeast. 2012 Dec;29(12):495-503. doi: 10.1002/yea.2930. Epub 2012 Nov 19. Yeast. 2012. PMID: 23166051 - Using regulatory information to manipulate glycerol metabolism in Saccharomyces cerevisiae.
Hou J, Vemuri GN. Hou J, et al. Appl Microbiol Biotechnol. 2010 Jan;85(4):1123-30. doi: 10.1007/s00253-009-2202-6. Epub 2009 Sep 2. Appl Microbiol Biotechnol. 2010. PMID: 19727706 - Gpd1 and Gpd2 fine-tuning for sustainable reduction of glycerol formation in Saccharomyces cerevisiae.
Hubmann G, Guillouet S, Nevoigt E. Hubmann G, et al. Appl Environ Microbiol. 2011 Sep;77(17):5857-67. doi: 10.1128/AEM.05338-11. Epub 2011 Jul 1. Appl Environ Microbiol. 2011. PMID: 21724879 Free PMC article. - Natural and modified promoters for tailored metabolic engineering of the yeast Saccharomyces cerevisiae.
Hubmann G, Thevelein JM, Nevoigt E. Hubmann G, et al. Methods Mol Biol. 2014;1152:17-42. doi: 10.1007/978-1-4939-0563-8_2. Methods Mol Biol. 2014. PMID: 24744025 Review. - Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae.
He S, Zhang Z, Lu W. He S, et al. J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuac029. doi: 10.1093/jimb/kuac029. J Ind Microbiol Biotechnol. 2023. PMID: 36633543 Free PMC article. Review.
Cited by
- Production and quantification of sesquiterpenes in Saccharomyces cerevisiae, including extraction, detection and quantification of terpene products and key related metabolites.
Rodriguez S, Kirby J, Denby CM, Keasling JD. Rodriguez S, et al. Nat Protoc. 2014 Aug;9(8):1980-96. doi: 10.1038/nprot.2014.132. Epub 2014 Jul 24. Nat Protoc. 2014. PMID: 25058645 - One-Step Biosynthesis of Vitamin C in Saccharomyces cerevisiae.
Zhou M, Bi Y, Ding M, Yuan Y. Zhou M, et al. Front Microbiol. 2021 Feb 25;12:643472. doi: 10.3389/fmicb.2021.643472. eCollection 2021. Front Microbiol. 2021. PMID: 33717042 Free PMC article. - Promoter Architecture and Promoter Engineering in Saccharomyces cerevisiae.
Tang H, Wu Y, Deng J, Chen N, Zheng Z, Wei Y, Luo X, Keasling JD. Tang H, et al. Metabolites. 2020 Aug 6;10(8):320. doi: 10.3390/metabo10080320. Metabolites. 2020. PMID: 32781665 Free PMC article. Review. - Combining systems and synthetic biology for in vivo enzymology.
Castaño-Cerezo S, Chamas A, Kulyk H, Treitz C, Bellvert F, Tholey A, Galéote V, Camarasa C, Heux S, Garcia-Alles LF, Millard P, Truan G. Castaño-Cerezo S, et al. EMBO J. 2024 Nov;43(21):5169-5185. doi: 10.1038/s44318-024-00251-w. Epub 2024 Sep 25. EMBO J. 2024. PMID: 39322757 Free PMC article. - Novel S. cerevisiae Hybrid Synthetic Promoters Based on Foreign Core Promoter Sequences.
Feng X, Marchisio MA. Feng X, et al. Int J Mol Sci. 2021 May 27;22(11):5704. doi: 10.3390/ijms22115704. Int J Mol Sci. 2021. PMID: 34071849 Free PMC article.
References
- Albertyn, J., S. Hohmann, J. M. Thevelein, and B. A. Prior. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14:4135-4144. - PMC - PubMed
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous