Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae - PubMed (original) (raw)
Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae
Robert M Seiser et al. Genetics. 2006 Oct.
Abstract
In eukaryotes, 40S and 60S ribosomal subunits are assembled in the nucleus and exported to the cytoplasm independently of one another. Nuclear export of the 60S requires the adapter protein Nmd3, but no analogous adapter has been identified for the 40S. Ltv1 is a nonessential, nonribosomal protein that is required for 40S subunit biogenesis in yeast. Cells lacking LTV1 grow slowly, are hypersensitive to inhibitors of protein synthesis, and produce about half as many 40S subunits as do wild-type cells. Ltv1 interacts with Crm1, co-sediments in sucrose gradients with 43S/40S subunits, and copurifies with late 43S particles. Here we show that Ltv1 shuttles between nucleus and cytoplasm in a Crm1-dependent manner and that it contains a functional NES that is sufficient to direct the export of an NLS-containing reporter. Small subunit export is reduced in Deltaltv1 mutants, as judged by the altered distribution of the 5'-ITS1 rRNA and the 40S ribosomal protein RpS3. Finally, we show a genetic interaction between LTV1 and YRB2, a gene that encodes a Ran-GTP-, Crm1-binding protein that facilitates the small subunit export. We propose that Ltv1 functions as one of several possible adapter proteins that link the nuclear export machinery to the small subunit.
Figures
Figure 1.—
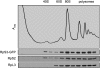
Ribosomal RNA-processing defects in Δ_yar1_ and Δ_ltv1_ strains. Wild-type parental strain (LY134) and isogenic deletion strains Δ_yar1_ (LY135) and Δ_ltv1_ (LY136) were grown at 30° in YPD medium. Following total RNA extraction, 10 μg of RNA from each strain was subjected to formaldehyde–agarose gel electrophoresis and processed for Northern blotting. Blots were probed with the 32P-labeled oligonucleotide probes depicted at the top of the figure (adapted from K
ressler
et al. 1997). The sizes of the various rRNA intermediate species are indicated. A schematic of the structure of these intermediates is shown at the right.
Figure 2.—
p_LTV1_-GFP rescues the Δ_ltv1_ cold-sensitive mutant phenotype. Wild-type (WT) (LY134) and Δ_ltv1_ (LY136) strains transformed with either pGFP (pUG23) or p_LTV1_-GFP (Ld47) were grown in selective media to midlog phase. Cell density was determined by hemocytometer and cells were diluted to 3.2 × 106 cells/ml. Serial 1:4 dilutions were made and 5-μl aliquots of each dilution were spotted onto plates lacking histidine and containing 500 μ
m
methionine. Plates were incubated at 30° and photographed after 37 hr or at 18° and photographed after 64 hr.
Figure 3.—
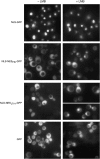
Nuclear export of Ltv1-GFP is Crm1 dependent. (A) p_LTV1_-GFP (Ld47), p_YAR1_-GFP (Ld46), and NLSSV40-NESPKI-GFP (pPS1372) plasmids were transformed in the LMB-sensitive crm1(T539C) (MNY8) strain. Strains expressing these GFP-tagged proteins were grown overnight at 30° in appropriate selective media to early to midlog phase, harvested, and photographed as described in
materials and methods
. LMB was added to an aliquot of each culture to a final concentration of 100 ng/ml and images of living cells captured beginning 5 and 60 min after addition of LMB. Time, in the left-most column, is the time since addition of LMB to cells. (B) Living cells expressing Ltv1-GFP (Ld47) or NLSSV40-NESPKI-GFP (pPS1372) protein were stained with DAPI and then treated with LMB. Cells were photographed 5 min after the addition of LMB.
Figure 4.—
Sequence alignment identifies a potential NES in Ltv1. Ten experimentally validated NES sequences were aligned in Clustal X. Profile alignment was then used to align this NES profile to the S. cerevisiae Ltv1 sequence. Only one sequence was identified in Ltv1, which was then compared to the aligned fungal Ltv1 proteins to determine whether this sequence was conserved between distantly related fungal species. The conserved sequence is shown here.
Figure 5.—
Ltv1 NES is sufficient to drive nuclear export of NLS-GFP reporter. MNY8 cells, transformed with pNLS-GFP (pPS815), pNLS-NESPKI-GFP (pPS1372), pNLS-NES LTV1-GFP (Ld61), or pGFP (pUG23), were grown in selective media to early log phase, collected, and washed in sterile water. Images of cells were captured using fluorescent microscopy and then LMB was added to an aliquot of cells to a final concentration of 100 ng/ml and photographed at 15 min after addition.
Figure 6.—
A fluorescent-tagged RpS3 fusion protein is incorporated into functional ribosomes. Wild-type (LY134) cells transformed with p_RPS3_-GFP (Ld50) were cultured to early log phase in media lacking histidine and methionine and then fractionated to yield a postnuclear homogenate. Ribosomal complexes were further separated on a 15–50% sucrose gradient. Individual gradient fractions were processed for Western blotting with antibodies to GFP and to small (S2) and large (L3) ribosomal subunit proteins.
Figure 7.—
RpS3-GFP export is retarded in Δ_ltv1_ cells. Wild-type (WT) (LY134) and Δ_ltv1_ (LY136) cells transformed with either p_RPS3_-GFP (Ld50) or pGFP (pUG23) were grown in media lacking histidine and methionine to early log phase, harvested, and visualized by fluorescence microscopy as described in
materials and methods
. Arrows identify the nuclear-excluded phenotype scored in Table 3.
Figure 8.—
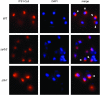
Deletion of YRB2 or LTV1 results in altered localization of an ITS1 rRNA fragment. A Cy3-labeled DNA probe complementary to the 5′-ITS1 sequence was hybridized with fixed and permeabilized wild-type (WT) (BY4743), Δ_yrb2_ (PSY2070), or Δ_ltv1_ (LY139) mutant cells. The nuclear DNA was counterstained with DAPI prior to viewing in an epifluorescence microscope. Overlaid images show the colocalization or apposition of the ITS1 and DAPI signals. Cells exhibiting distinct, nonoverlapping DAPI/ITS1 signals, as scored in Table 4, are indicated with arrowheads.
Figure 9.—
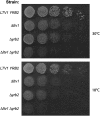
Genetic interaction between LTV1 and YRB2. (A) Wild-type strain LY134 (BY4742), its isogenic derivative LY136 (Δ_ltv1∷KanR_), PSY2070 (Δ_yrb2∷HIS3_), and the double-mutant LY181 (Δ_yrb2∷HIS3_ Δ_ltv1∷KanR_) were grown to midlog phase in YPD liquid media. Cell density was determined by hemocytometer and each culture was diluted to 3.2 × 106 cells/ml and then serially diluted 1:4 four times. We spotted 5-μl aliquots of each dilution from left to right on two YPD plates, which were incubated at 18° (photographed after 4 days) or at 30° (photographed after 1.5 days).
Figure 10.—
Toxicity of Yrb2 overexpression requires Ltv1. Wild-type (LY134) and Δ_ltv1_ (LY136) strains transformed either with the empty vector (pGAL) or with pPS1082 (pGAL-YRB2) were grown in selective media containing 2% glucose or 2% galactose to midlog phase. Cells were diluted and spotted onto selective plates containing the same galactose/glucose concentrations as the liquid media and incubated as described in Figure 2.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis.
Dong J, Lai R, Jennings JL, Link AJ, Hinnebusch AG. Dong J, et al. Mol Cell Biol. 2005 Nov;25(22):9859-73. doi: 10.1128/MCB.25.22.9859-9873.2005. Mol Cell Biol. 2005. PMID: 16260602 Free PMC article. - The C-terminal tail of ribosomal protein Rps15 is engaged in cytoplasmic pre-40S maturation.
Rössler I, Weigl S, Fernández-Fernández J, Martín-Villanueva S, Strauss D, Hurt E, de la Cruz J, Pertschy B. Rössler I, et al. RNA Biol. 2022;19(1):560-574. doi: 10.1080/15476286.2022.2064073. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35438042 Free PMC article. - Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis.
Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. Rosado IV, et al. Mol Cell Biol. 2007 Feb;27(4):1207-21. doi: 10.1128/MCB.01523-06. Epub 2006 Dec 4. Mol Cell Biol. 2007. PMID: 17145778 Free PMC article. - Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.
Triana L, Palacios Huatuco RM, Campilgio G, Liscano E. Triana L, et al. Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
- Location is everything: an educational primer for use with "genetic analysis of the ribosome biogenesis factor Ltv1 of Saccharomyces cerevisiae".
Edwalds-Gilbert G. Edwalds-Gilbert G. Genetics. 2015 Feb;199(2):307-13. doi: 10.1534/genetics.114.173641. Genetics. 2015. PMID: 25657348 Free PMC article. - Hrr25/CK1δ-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth.
Ghalei H, Schaub FX, Doherty JR, Noguchi Y, Roush WR, Cleveland JL, Stroupe ME, Karbstein K. Ghalei H, et al. J Cell Biol. 2015 Mar 16;208(6):745-59. doi: 10.1083/jcb.201409056. J Cell Biol. 2015. PMID: 25778921 Free PMC article. - A general method for quantitative fractionation of mammalian cells.
Udi Y, Zhang W, Stein ME, Ricardo-Lax I, Pasolli HA, Chait BT, Rout MP. Udi Y, et al. J Cell Biol. 2023 Jun 5;222(6):e202209062. doi: 10.1083/jcb.202209062. Epub 2023 Mar 15. J Cell Biol. 2023. PMID: 36920247 Free PMC article. - Role of Mex67-Mtr2 in the nuclear export of 40S pre-ribosomes.
Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG. Faza MB, et al. PLoS Genet. 2012;8(8):e1002915. doi: 10.1371/journal.pgen.1002915. Epub 2012 Aug 30. PLoS Genet. 2012. PMID: 22956913 Free PMC article. - The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Cheng J, Lau B, Thoms M, Ameismeier M, Berninghausen O, Hurt E, Beckmann R. Cheng J, et al. Nucleic Acids Res. 2022 Nov 11;50(20):11924-11937. doi: 10.1093/nar/gkac961. Nucleic Acids Res. 2022. PMID: 36321656 Free PMC article.
References
- Amberg, D. C., A. L. Goldstein and C. N. Cole, 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6: 1173–1189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM-061643/GM/NIGMS NIH HHS/United States
- F32 GM068208/GM/NIGMS NIH HHS/United States
- GM-068208/GM/NIGMS NIH HHS/United States
- R15 GM061643/GM/NIGMS NIH HHS/United States
- GM-061643-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous