TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients - PubMed (original) (raw)
. 2006 Aug 23;25(16):3832-42.
doi: 10.1038/sj.emboj.7601262. Epub 2006 Aug 3.
Aiko Kitamura, Ryo Shioda, Hironori Maruyama, Kanako Ozaki, Yoko Oda, Thierry Mini, Paul Jenö, Yasushi Maki, Kazuyoshi Yonezawa, Ed Hurt, Masaru Ueno, Masahiro Uritani, Michael N Hall, Takashi Ushimaru
Affiliations
- PMID: 16888624
- PMCID: PMC1553199
- DOI: 10.1038/sj.emboj.7601262
TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients
Yoshimi Honma et al. EMBO J. 2006.
Abstract
The protein kinase TOR (target of rapamycin) controls several steps of ribosome biogenesis, including gene expression of rRNA and ribosomal proteins, and processing of the 35S rRNA precursor, in the budding yeast Saccharomyces cerevisiae. Here we show that TOR also regulates late stages of ribosome maturation in the nucleoplasm via the nuclear GTP-binding protein Nog1. Nog1 formed a complex that included 60S ribosomal proteins and pre-ribosomal proteins Nop7 and Rlp24. The Nog1 complex shuttled between the nucleolus and the nucleoplasm for ribosome biogenesis, but it was tethered to the nucleolus by both nutrient depletion and TOR inactivation, causing cessation of the late stages of ribosome biogenesis. Furthermore, after this, Nog1 and Nop7 proteins were lost, leading to complete cessation of ribosome maturation. Thus, the Nog1 complex is a critical regulator of ribosome biogenesis mediated by TOR. This is the first description of a physiological regulation of nucleolus-to-nucleoplasm translocation of pre-ribosome complexes.
Figures
Figure 1
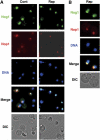
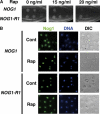
TOR inactivation causes Nog1 accumulation in the nucleolus. (A) Nog1 is accumulated in the nucleolus upon rapamycin treatment. Cells of a strain expressing NOG1-GFP (SCU888) with plasmid pSCU632 (pDsRed-NOP1) were treated with rapamycin (200 ng/ml) for 30 min (Rap). GFP (green), DsRed (red) and DAPI (blue) signals were observed in cells. Nomarski images (DIC) were also recorded. Cells not treated with rapamycin were used as the control (Cont). (B) Enlarged images of rapamycin-treated cells.
Figure 2
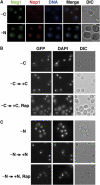
Nutrient starvation causes Nog1 accumulation in the nucleolus. (A) Cells of a strain expressing NOG1-GFP (SCU888) with plasmid pSCU632 (pDsRed-NOP1) were transferred to carbon- (−C) or nitrogen-deprived (−N) medium for 30 min (−C). GFP (green), DsRed (red) and DAPI (blue) signals were observed in cells. (B) Nog1 is accumulated in the nucleolus upon carbon starvation. Cells of a strain expressing NOG1-GFP (SCU888) were transferred to carbon-deprived medium for 30 min (−C) and then re-supplemented with carbon in the presence (−C → +C, Rap) or absence (−C → +C) of rapamycin for 60 min. GFP and DAPI signals were observed in cells. (C) Nog1 is accumulated in the nucleolus upon nitrogen starvation. The same strain was transferred to nitrogen-deprived medium for 30 min (−N) and then re-supplemented with nitrogen in the presence (−N → +N, Rap) or absence (−N → +N) of rapamycin for 60 min. GFP and DAPI signals were observed in cells.
Figure 3
Pre-60S complex was accumulated in the nucleus as a result of Nog1 dysfunction. (A) Temperature-sensitive (ts) growth phenotype of nog1-ts mutants. The wild-type NOG1 (SCU801) and nog1 temperature-sensitive nog1-11 (SCU823), nog1-12 (SCU824) and nog1-13 (SCU825) cells were incubated on plates at 25°C for 3 days or at 37°C for 2 days. (B) Pre-60S complex was accumulated in the nucleus of nog1 temperature-sensitive mutants. Strains NOG1 (SCU783), nog1-11 (SCU823), nog1-12 (SCU824) and nog1-13 (SCU825), which harbor a plasmid expressing GFP-Rpl25 (pSCU615), were incubated at 37°C for 8 h and GFP signals were observed. (C) Strain nog1-11 (SCU823) harboring plasmid pGFP-RPL25 (pSCU615) was incubated at 37°C for 5 h and stained for GFP (Rpl25; green) and DAPI (blue).
Figure 4
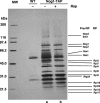
Identification of components of the Nog1 complex. Cells of a strain expressing NOG1-TAP (SCU889) were harvested before (lane a) and 30 min after rapamycin (200 ng/ml) treatment (lane b). Nog1-associated proteins extracted from cells were purified according to the TAP purification protocol, and were separated by SDS–PAGE followed by silver staining. Wild-type strain TB50a (SCU425) was used as the negative control. Ribosomal (RP) and pre-ribosomal (Pre-RP) proteins were identified by mass spectrometric analysis.
Figure 5
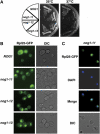
Nop7 functions together with Nog1. (A) Overexpression of NOP7 suppresses thermosensitivity of nog1-ts strains. Strains nog1-11 (SCU823), nog1-13 (SCU825) and the wild-type NOG1 (SCU822) were transformed with a plasmid for overexpression of NOP7 (pSCU674) or with the empty vector (pSCU154). Serially diluted cells of each strain were spotted from the left to the right on YPAD plates. Plates were cultured at 25°C for 2 days or at 37°C for 1 day. (B) Overexpression of NOG1 suppresses thermosensitivity of nop7-td strains. nop7-td (SCU89) and the wild-type NOP7 (SCU88) strains were transformed with a plasmid for overexpression of NOG1 (pSCU388) or with the empty vector (pSCU379). Serially diluted cells of each strain were spotted from left to right on YPAD plates. Plates were cultured at 25°C for 2 days or at 34°C for 1 day. (C) Nop7 shows the STING phenotype. Cells of a strain expressing NOP7-GFP (SCU888) harboring a plasmid expressing DsRed-NOP1 (pSCU618) were treated with rapamycin (200 ng/ml, Rap) for 60 min. GFP (Nop7, green), RFP (Nop1, red) and DAPI (blue) signals were observed. Nontreated cells were used as the control (Cont).
Figure 6
Effects of rapamycin on localization of Rpl24 and Has1. (A) Strain SCU7 harboring a plasmid expressing RPL24-Myc13GFP (pSCU771) and DsRed-NOP1 (pSCU618) was treated with rapamycin (200 ng/ml, Rap) for 60 min, and GFP (Rlp24, green), RFP (Nop1, red) and DAPI (blue) signals were examined. Nontreated cells were used as the control (Cont). (B) Cells expressing HA2-HAS1 (SCU1285) were treated with rapamycin (200 ng/ml, Rap) for 60 min and stained for HA (Has1; green), Nop1 (red) and DAPI (blue).
Figure 7
Rapamycin-resistant NOG1-R1 strain is defective in STING. (A) Rapamycin-resistant growth of the NOG1-R1 mutant strain. Serially diluted cells of the wild-type NOG1 (SCU801) and NOG1-R1 (SCU1287) strains were spotted from left to right on YPAD plates containing various concentrations of rapamycin at 30°C for 3 days. (B) NOG1-R1 cells are defective in STING. NOG1 (SCU801) and NOG1-R1 (SCU1287) strains were treated with 50 ng/ml rapamycin for 60 min. Both types of cells expressed Nog1-HA3 and were stained for HA (Nog1; green) and DAPI (blue).
Figure 8
TOR regulates concentration of the Nog1 complex proteins. (A) NOG1 expression is upregulated by TOR. Cells (SCU7) were treated with rapamycin (200 ng/ml) or transferred to medium without a nitrogen or carbon source for 15 min. ACT1 was used as the loading control. (B) Changes in protein levels of Nog1, Nop7 and Nop1 after rapamycin treatment. NOP7-myc13 strain (SCU799) with plasmid pHAC33-NOG1 (pSCU387) was treated with rapamycin (200 ng/ml). Whole-cell extracts were subjected to Western blotting using anti-HA, anti-Myc, anti-Nop1 and anti-cyclin-dependent kinase (Cdk) antibodies. Cdk was used as the loading control. (C) Changes in protein levels of Nog1 after shut-off of NOG1 expression in the presence or absence of rapamycin. The wild-type cells (SCU425) harboring plasmid pGAL-HA-NOG1 (pSCU679) were pre-cultured in galactose-based medium for expressing HA-NOG1 and then supplemented with glucose for shut-off of NOG1 expression (at time 0). Rapamycin (200 ng/ml) was added simultaneously to cultures (+Rap). Cells not treated with rapamycin were prepared as the control (−Rap).
Figure 9
Rapamycin sensitivities and Nog1 protein levels of nog1-ts mutants. (A) Rapamycin-sensitive growth of nog1-11 and nog1-12. Serially diluted cells of NOG1 (SCU801), nog1-11, nog1-12 and nog1-13 strains were spotted from left to right on YPAD plates containing various concentrations of rapamycin at 30°C for 3 days. (B) Nog1 protein level of nog1-ts is decreased after shift to restrictive temperature in the presence of rapamycin. Each nog1-ts strain grown at 25°C was shifted to 37°C in the presence or absence of rapamycin (200 ng/ml) and the amount of Nog1 protein was determined by Western blotting using anti-HA antibody.
Similar articles
- Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes.
Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, Ma C, Lei J, Dong MQ, Woolford JL Jr, Gao N. Wu S, et al. Nature. 2016 Jun 2;534(7605):133-7. doi: 10.1038/nature17942. Epub 2016 May 25. Nature. 2016. PMID: 27251291 Free PMC article. - The GTPase Nog1 co-ordinates the assembly, maturation and quality control of distant ribosomal functional centers.
Klingauf-Nerurkar P, Gillet LC, Portugal-Calisto D, Oborská-Oplová M, Jäger M, Schubert OT, Pisano A, Peña C, Rao S, Altvater M, Chang Y, Aebersold R, Panse VG. Klingauf-Nerurkar P, et al. Elife. 2020 Jan 7;9:e52474. doi: 10.7554/eLife.52474. Elife. 2020. PMID: 31909713 Free PMC article. - TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre-40S ribosome export.
Vanrobays E, Leplus A, Osheim YN, Beyer AL, Wacheul L, Lafontaine DL. Vanrobays E, et al. RNA. 2008 Oct;14(10):2061-73. doi: 10.1261/rna.1176708. Epub 2008 Aug 28. RNA. 2008. PMID: 18755838 Free PMC article. - Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.
Konikkat S, Woolford JL Jr. Konikkat S, et al. Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review. - The nucleolus: an organelle formed by the act of building a ribosome.
Mélèse T, Xue Z. Mélèse T, et al. Curr Opin Cell Biol. 1995 Jun;7(3):319-24. doi: 10.1016/0955-0674(95)80085-9. Curr Opin Cell Biol. 1995. PMID: 7662360 Review.
Cited by
- Prasinovirus Attack of Ostreococcus Is Furtive by Day but Savage by Night.
Derelle E, Yau S, Moreau H, Grimsley NH. Derelle E, et al. J Virol. 2018 Jan 30;92(4):e01703-17. doi: 10.1128/JVI.01703-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187539 Free PMC article. - In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1.
Fuentes JL, Datta K, Sullivan SM, Walker A, Maddock JR. Fuentes JL, et al. Mol Genet Genomics. 2007 Jul;278(1):105-23. doi: 10.1007/s00438-007-0233-1. Epub 2007 Apr 19. Mol Genet Genomics. 2007. PMID: 17443350 - Concerted modification of nucleotides at functional centers of the ribosome revealed by single-molecule RNA modification profiling.
Bailey AD, Talkish J, Ding H, Igel H, Duran A, Mantripragada S, Paten B, Ares M. Bailey AD, et al. Elife. 2022 Apr 6;11:e76562. doi: 10.7554/eLife.76562. Elife. 2022. PMID: 35384842 Free PMC article. - Nucleolar GTPase NOG-1 regulates development, fat storage, and longevity through insulin/IGF signaling in C. elegans.
Kim YI, Bandyopadhyay J, Cho I, Lee J, Park DH, Cho JH. Kim YI, et al. Mol Cells. 2014 Jan;37(1):51-7. doi: 10.14348/molcells.2014.2251. Epub 2014 Jan 27. Mol Cells. 2014. PMID: 24552710 Free PMC article. - Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae.
Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL Jr. Talkish J, et al. Nucleic Acids Res. 2012 Sep 1;40(17):8646-61. doi: 10.1093/nar/gks609. Epub 2012 Jun 26. Nucleic Acids Res. 2012. PMID: 22735702 Free PMC article.
References
- Allen NP, Huang L, Burlingame A, Rexach M (2001) Proteomic analysis of nucleoporin interacting proteins. J Biol Chem 276: 29268–29274 - PubMed
- Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ (2004) Phosphorylation by cyclin B–Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305: 516–519 - PubMed
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E (2001) Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases