Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling - PubMed (original) (raw)
Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling
Shih-Ching Lo et al. EMBO J. 2006.
Abstract
Keap1 is a BTB-Kelch substrate adaptor protein that regulates steady-state levels of Nrf2, a bZIP transcription factor, in response to oxidative stress. We have determined the structure of the Kelch domain of Keap1 bound to a 16-mer peptide from Nrf2 containing a highly conserved DxETGE motif. The Nrf2 peptide contains two short antiparallel beta-strands connected by two overlapping type I beta-turns stabilized by the aspartate and threonine residues. The beta-turn region fits into a binding pocket on the top face of the Kelch domain and the glutamate residues form multiple hydrogen bonds with highly conserved residues in Keap1. Mutagenesis experiments confirmed the role of individual amino acids for binding of Nrf2 to Keap1 and for Keap1-mediated repression of Nrf2-dependent gene expression. Our results provide a detailed picture of how a BTB-Kelch substrate adaptor protein binds to its cognate substrate and will enable the rational design of novel chemopreventive agents.
Figures
Figure 1
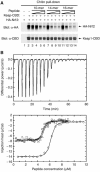
(A) The sequence of the Kelch domains from the Keap1 protein of five different species (human, mouse, zebrafish, Drosophila melanogaster, and Anopheles gambia) is shown, along with the sequence of three human BTB-Kelch proteins, mayven, gigaxonin, and ENC1. The six blades of the β-propeller structure are indicated above the alignment and the four β-strands (A–D) that comprise each blade are highlighted in gray. The A strand for blade I is located at the extreme C-terminus of the protein. Disease-associated mutations in Keap1, gigaxonin, and ENC1 proteins are underlined in bold (Bomont et al, 2000, 2003; Bruno et al, 2004; Kuhlenbaumer et al, 2002; Liang et al, 2004; Padmanabhan et al, 2006). Highly conserved residues that define the Kelch repeat are indicated by asterisks. Amino acids in Keap1 whose side chains contact the Nrf2-derived peptide in the crystal structure are highlighted in blue, whereas those amino acids that only make van der Waals contacts with the Nrf2-derived peptide are highlighted in green. (B) The Neh2 domain from the human Nrf1 and Nrf2 proteins is shown, along with the corresponding regions of Nrf2-related proteins from zebrafish and D. melanogaster. Two conserved motifs that have been implicated in binding to Keap1 are shown. The glutamate residues of the DxETGE motif are highlighted in blue. The sequence of the three Nrf2-derived peptides used in our experiments is shown.
Figure 2
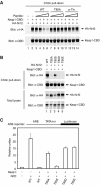
(A) COS1 cells were transfected with expression vectors for Keap1-CBD and HA-Nrf2 and the Keap1:Nrf2 complex isolated on chitin beads. The beads were incubated with the indicated peptides, washed, and proteins that remained bound to the beads were analyzed by immunoblot with anti-HA and anti-CBD antibodies. The amounts of peptides added to each sample were 10 ng (lanes 3, 7, 11), 100 ng (lanes 4, 8, 12), 1 μg (lanes 5, 9, 13), and 10 μg (lanes 6, 10, 14). (B) (Upper panel) Thermographs from a representative isothermal calorimetry experiment are shown, in which a 5 μM solution of the Kelch domain was titrated with 50 μM peptide. (Lower panel) The fitted binding isotherms from four experiments are shown. Three experiments (circles, squares, and triangles) were performed by titrating a 50 μM peptide solution into a 5 μM solution of the Kelch domain. A fourth experiment (diamonds) was performed by titrating an 88 μM solution of the peptide into a 6.25 μM solution of the Kelch domain.
Figure 3
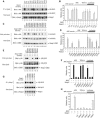
(A) A ribbon diagram of the Kelch β-propeller (red) and bound Nrf2 peptide (yellow tube). The termini of the peptide are labeled N-ter and C-ter; those of the Kelch domain are labeled N and C. The six blades of the β-propeller are labeled I–VI and the four β-strands found in each blade are labeled A–D (white font) on blade VI. (B) Side view of the Kelch domain, with a surface representation of the β-propeller. (C) A surface representation of the Kelch propeller (gray) and peptide (yellow tube). Selected residues are shown in blue (basic), orange (polar), and green (apolar).
Figure 4
(A) Contacts between the side chain atoms of the Nrf2 peptide and residues in the Kelch domain. (B) Contacts between the backbone atoms of the peptide and residues of the Kelch domain. (C) A stick model of the Nrf2 peptide, with intramolecular hydrogen bonds highlighted. An _F_o−_F_c electron density omit map contoured at 2.5σ for the vicinity of the peptide is shown in blue. Phases for the map were determined immediately following molecular replacement, before the inclusion of the peptide in the model. The side chain of E78 was not well ordered in the electron density maps and is shown as semi-transparent. (D) A schematic showing both backbone and side-chain contacts to the Nrf2 peptide (yellow/blue) from interacting residues in the Kelch domain.
Figure 5
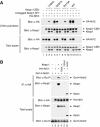
(A) COS1 cells were cotransfected with expression vectors for HA-Nrf2 and mutant Keap1 proteins as indicated. Total cell lysates were analyzed by immunoblot with anti-HA and anti-Keap1 antibodies (bottom two panels). Anti-Keap1 immunoprecipitates (IP) were subjected to immunoblot analysis using anti-HA antibodies (top panel). (B) MDA-MB-231 cells were transfected with expression vectors for HA-Nrf2 (100 ng) and mutant Keap1 proteins (50 ng) as indicated, and with an ARE-dependent firefly luciferase reporter gene construct (100 ng). A plasmid encoding Renilla luciferase (10 ng) was included as a control for transfection efficiency. The data shown represent the means and standard deviation of results from three independent experiments. (C) HEK 293 T cells were transfected with expression vectors for Keap1-CBD and mutant HA-Nrf2 proteins as indicated. Total cell lysates were analyzed by immunoblotting with anti-HA and anti-CBD antibodies (bottom two panels). The lysates were incubated with chitin beads, washed, and proteins that remained associated with the chitin beads were analyzed by immunoblotting with anti-HA antibodies (top panel). An asterisk (*) indicates a nonspecific protein detected by the antibody. (D) Reporter assays were formed as described in (B). (E) Pulldown assays were performed as described for (C). (F) Reporter assays were performed as described in (B). (G) Co-immunoprecipitation assays were performed as described in (A), except that the Keap1 and Nrf2 expression vectors were separately transfected into cells and cell lysates were mixed before the immunoprecipitation after input amounts were normalized to Nrf2 levels. (H). Reporter assays were performed as described in (B).
Figure 6
(A) HEK 293 T cells were transfected with expression vectors for the indicated Keap1 and Nrf2 proteins. Total cell lysates were analyzed by immunoblotting with anti-Keap1 and anti-HA antibodies (bottom two panels). Equivalent amounts of cell lysates were incubated with chitin beads, washed, and proteins that remained associated with the chitin beads were analyzed by immunoblotting with anti-HA and anti-Keap1 antibodies (top two panels). (B) HEK 293 T cells were transfected with expression vectors for the indicated Keap1 and Nrf2 proteins. Total cell lysates were analyzed by immunoblotting with anti-HA, anti-Keap1, and anti-Gal4 antibodies (bottom three panels). Anti-HA immunoprecipitates were analyzed by immunoblotting with anti-Gal4 and anti-Keap1 antibodies (top two panels).
Figure 7
(A) COS1 cells were transfected with expression vectors for Keap1-CBD and HA-Nrf2 and the Keap1:Nrf2 complex isolated on chitin beads. The beads were incubated with the indicated peptides, washed, and proteins that remained bound to the beads were analyzed by immunoblot with anti-HA and anti-CBD antibodies. The amounts of peptides added to each sample were 10 ng (lanes 3, 7, 11), 100 ng (lanes 4, 8, 12), 1 μg (lanes 5, 9, 13), and 10 μg (lanes 6, 10, 14). (B) HEK 293 T cells were transfected with expression vectors for the indicated Keap1 and Nrf2 proteins. Total cell lysates were analyzed by immunoblotting with anti-HA and anti-CBD antibodies (bottom two panels). Equivalent amounts of cell lysates were incubated with chitin beads, washed, and proteins that remained associated with the chitin beads were analyzed by immunoblotting with anti-HA and anti-CBD antibodies (top two panels). (C) Reporter assays were performed as described in Figure 5B.
Similar articles
- Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum.
Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Zhang Y, et al. Biochem J. 2006 Nov 1;399(3):373-85. doi: 10.1042/BJ20060725. Biochem J. 2006. PMID: 16872277 Free PMC article. - Identification and characterization of novel Nrf2 inducers designed to target the intervening region of Keap1.
Wu JH, Miao W, Hu LG, Batist G. Wu JH, et al. Chem Biol Drug Des. 2010 May;75(5):475-80. doi: 10.1111/j.1747-0285.2010.00955.x. Chem Biol Drug Des. 2010. PMID: 20486933 - Structural insights into the multiple binding modes of Dimethyl Fumarate (DMF) and its analogs to the Kelch domain of Keap1.
Unni S, Deshmukh P, Krishnappa G, Kommu P, Padmanabhan B. Unni S, et al. FEBS J. 2021 Mar;288(5):1599-1613. doi: 10.1111/febs.15485. Epub 2020 Aug 7. FEBS J. 2021. PMID: 32672401 - Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome.
Katoh Y, Iida K, Kang MI, Kobayashi A, Mizukami M, Tong KI, McMahon M, Hayes JD, Itoh K, Yamamoto M. Katoh Y, et al. Arch Biochem Biophys. 2005 Jan 15;433(2):342-50. doi: 10.1016/j.abb.2004.10.012. Arch Biochem Biophys. 2005. PMID: 15581590 Review. - Structural basis of Keap1 interactions with Nrf2.
Canning P, Sorrell FJ, Bullock AN. Canning P, et al. Free Radic Biol Med. 2015 Nov;88(Pt B):101-107. doi: 10.1016/j.freeradbiomed.2015.05.034. Epub 2015 Jun 7. Free Radic Biol Med. 2015. PMID: 26057936 Free PMC article. Review.
Cited by
- The Role of NRF2/KEAP1 Signaling Pathway in Cancer Metabolism.
Song MY, Lee DY, Chun KS, Kim EH. Song MY, et al. Int J Mol Sci. 2021 Apr 22;22(9):4376. doi: 10.3390/ijms22094376. Int J Mol Sci. 2021. PMID: 33922165 Free PMC article. Review. - Cullin3-KLHL15 ubiquitin ligase mediates CtIP protein turnover to fine-tune DNA-end resection.
Ferretti LP, Himmels SF, Trenner A, Walker C, von Aesch C, Eggenschwiler A, Murina O, Enchev RI, Peter M, Freire R, Porro A, Sartori AA. Ferretti LP, et al. Nat Commun. 2016 Aug 26;7:12628. doi: 10.1038/ncomms12628. Nat Commun. 2016. PMID: 27561354 Free PMC article. - p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription.
Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T. Jain A, et al. J Biol Chem. 2010 Jul 16;285(29):22576-91. doi: 10.1074/jbc.M110.118976. Epub 2010 May 7. J Biol Chem. 2010. PMID: 20452972 Free PMC article. - Role of Nuclear Factor Erythroid 2 (Nrf2) in the Recovery of Long COVID-19 Using Natural Antioxidants: A Systematic Review.
Muchtaridi M, Amirah SR, Harmonis JA, Ikram EHK. Muchtaridi M, et al. Antioxidants (Basel). 2022 Aug 10;11(8):1551. doi: 10.3390/antiox11081551. Antioxidants (Basel). 2022. PMID: 36009268 Free PMC article. Review. - Structural analysis of the complex of Keap1 with a prothymosin alpha peptide.
Padmanabhan B, Nakamura Y, Yokoyama S. Padmanabhan B, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Apr 1;64(Pt 4):233-8. doi: 10.1107/S1744309108004995. Epub 2008 Mar 21. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18391415 Free PMC article.
References
- Ames BN, Shigenaga MK (1993) DNA and free radicals. In Oxidants are a Major Contributor to Cancer and Aging, Halliwell B, Aruoma OI (eds) pp 1–15. New York, NY
- Bloom D, Dhakshinamoorthy S, Jaiswal AK (2002) Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene 21: 2191–2200 - PubMed
- Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tuysuz B, Landrieu P, Hentati F, Koenig M (2000) The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet 26: 370–374 - PubMed
- Bomont P, Ioos C, Yalcinkaya C, Korinthenberg R, Vallat JM, Assami S, Munnich A, Chabrol B, Kurlemann G, Tazir M, Koenig M (2003) Identification of seven novel mutations in the GAN gene. Hum Mutat 21: 446. - PubMed
- Bonnefont-Rousselot D (2002) Glucose and reactive oxygen species. Curr Opin Clin Nutr Metab Care 5: 561–568 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous