Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange - PubMed (original) (raw)
Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange
Felipe Cortés-Ledesma et al. EMBO Rep. 2006 Sep.
Abstract
Molecular studies on double-strand break (DSB) repair in mitosis are usually performed with enzymatically induced DSBs, but spontaneous DSBs might arise because of replication failures, for example when replication encounters nicks. To study repair of replication-born DSBs, we defined a system in Saccharomyces cerevisiae for the induction of a site-specific single-strand break. We show that a 21-base pair (bp) HO site is cleaved at only one strand by the HO endonuclease, with the resulting nick being converted into a DSB by replication during the S phase. Repair of such replication-born DSBs occurs by sister-chromatid exchange (SCE). We provide molecular evidence that cohesins are required for repair of replication-born DSBs by SCE, as determined in smc3, scc1 and scc2 mutants, but not for other recombinational repair events. This work opens new perspectives to understand the importance of single-strand breaks as a source of recombination and the relevance of cohesion in the repair of replication-born DSBs.
Figures
Figure 1
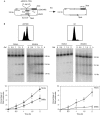
Breaks at the 21-bp HO site are primarily single-strand nicks. (A) Scheme of plasmid pRS316-TINV before and after the HO-induced DSB. Lines with asterisks indicate the LEU2 probe used for hybridization experiments. (B) Kinetics of HO cleavage as assayed by native and alkaline gel electrophoresis of DNA from asynchronous (ASYNC) and G1-arrested cultures of WS-bar1 strains. DNA samples were taken at the indicated time after HO induction and cut with _Xho_I and _Spe_I before electrophoresis. Fluorescence-activated cell sorting pattern of cultures and quantification data of nicks and DSBs are shown. The percentage of plasmids containing nicks was calculated as twice the difference between the number of broken molecules observed in the alkaline and native gels and normalized with respect to the total DNA fragments. Plasmids containing DSBs were determined directly from the native gels. DSBs, double-strand breaks; Gal, galactose.
Figure 2
Nicks are made in both strands. Breaks in each DNA strand were determined in alkaline gels by hybridization of DNA from G1-arrested cells with LEU2 single-stranded specific probes. For other details, see Fig 1. Gal, galactose.
Figure 3
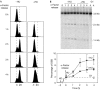
HO-induced double-strand breaks at the 21-bp HO site are formed after DNA replication. (A) Fluorescence-activated cell sorting analysis of the WS-bar1 cells arrested in G1 with α-factor and at different periods after release. (B) Kinetics of DSB formation (native gel) after release from G1 arrest, as determined by Southern blot analysis. After 2 h of HO induction in galactose (Gal), WS-bar1 G1-arrested cells were released from α-factor in the presence or absence of 50 mM HU. Quantification data are plotted at the bottom. For other details, see Fig 1. DSB, double-strand break; HU, hydroxyurea.
Figure 4
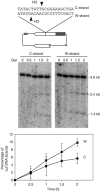
Cohesion is required for sister-chromatid exchange but not for intrachromatid recombination. (A) Schemes of plasmid pRS316-TINV and the intermediates produced by unequal SCE and ICR. Each band detected after _Xho_I–_Spe_I cleavage, using the LEU2 probe shown in Fig 1, is indicated with its corresponding size. Note that ICR events observed in these assays occur by break-induced replication, and gene conversions are not detected. (B) Kinetics of DSB formation, SCE and ICR intermediates in isogenic wild-type (wt) and smc3-42 strains. (C) Kinetics of DSB formation, SCE and ICR intermediates in isogenic scc1-73 and scc2-4 strains. Wt and smc3-73 data, taken from (B), are plotted for comparison. Cultures were incubated at restrictive temperature (37°C) for 30 min before HO induction. SCE levels were calculated as the ratio between the 4.7-kb band and the total plasmid DNA. ICR levels were calculated by subtracting the density value of the 4.7-kb band from that of the 2.9-kb band. For other details, see Fig 1. DSB, double-strand break; ICR, intrachromatid recombination; SCE, sister-chromatid exchange.
Figure 5
Cohesins are required for equal sister-chromatid exchange. (A) Schematic representation of the monomeric plasmid pCM189-leu2HOr, containing no DNA repeats, and the dimer obtained by equal SCE. (B) Kinetics of DSB formation and equal SCE intermediates in isogenic wild-type (wt) and smc3-42 strains. Plasmid DNA was not digested before electrophoresis. lM, linear monomers; rD, relaxed dimers; rM, relaxed monomers; scD, supercoiled dimers; scM, supercoiled monomers. Quantification of dimers (rD and scD) related to total plasmid DNA is shown. For other details, see Fig 1. DSB, double-strand break; SCE, sister-chromatid exchange.
Figure 6
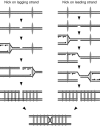
Model to explain the role of cohesins in the repair of double-strand breaks formed after replication forks encounter single-strand DNA breaks. Double-ended DSBs could be formed after replication through a nick in two ways depending on whether the nick is encountered at the lagging (left) or the leading strand (right). Cohesins (thin grey ovals), which are additionally loaded in the proximity of the DSB, favour HR with the sister. DSB, double-strand break; HR, homologous recombination.
Similar articles
- Rpd3L and Hda1 histone deacetylases facilitate repair of broken forks by promoting sister chromatid cohesion.
Ortega P, Gómez-González B, Aguilera A. Ortega P, et al. Nat Commun. 2019 Nov 15;10(1):5178. doi: 10.1038/s41467-019-13210-5. Nat Commun. 2019. PMID: 31729385 Free PMC article. - The Dot1 histone methyltransferase and the Rad9 checkpoint adaptor contribute to cohesin-dependent double-strand break repair by sister chromatid recombination in Saccharomyces cerevisiae.
Conde F, Refolio E, Cordón-Preciado V, Cortés-Ledesma F, Aragón L, Aguilera A, San-Segundo PA. Conde F, et al. Genetics. 2009 Jun;182(2):437-46. doi: 10.1534/genetics.109.101899. Epub 2009 Mar 30. Genetics. 2009. PMID: 19332880 Free PMC article. - Ctf18 is required for homologous recombination-mediated double-strand break repair.
Ogiwara H, Ohuchi T, Ui A, Tada S, Enomoto T, Seki M. Ogiwara H, et al. Nucleic Acids Res. 2007;35(15):4989-5000. doi: 10.1093/nar/gkm523. Epub 2007 Jul 18. Nucleic Acids Res. 2007. PMID: 17636314 Free PMC article. - S-phase and DNA damage activated establishment of sister chromatid cohesion--importance for DNA repair.
Sjögren C, Ström L. Sjögren C, et al. Exp Cell Res. 2010 May 15;316(9):1445-53. doi: 10.1016/j.yexcr.2009.12.018. Epub 2010 Jan 4. Exp Cell Res. 2010. PMID: 20043905 Review. - [Sister chromatid cohesion complex in Eukaryota].
Cena A, Kurlandzka A. Cena A, et al. Postepy Biochem. 2010;56(1):41-54. Postepy Biochem. 2010. PMID: 20499680 Review. Polish.
Cited by
- Induction of homologous recombination by site-specific replication stress.
Triplett MK, Johnson MJ, Symington LS. Triplett MK, et al. DNA Repair (Amst). 2024 Oct;142:103753. doi: 10.1016/j.dnarep.2024.103753. Epub 2024 Aug 16. DNA Repair (Amst). 2024. PMID: 39190984 Review. - PARG-deficient tumor cells have an increased dependence on EXO1/FEN1-mediated DNA repair.
Andronikou C, Burdova K, Dibitetto D, Lieftink C, Malzer E, Kuiken HJ, Gogola E, Ray Chaudhuri A, Beijersbergen RL, Hanzlikova H, Jonkers J, Rottenberg S. Andronikou C, et al. EMBO J. 2024 Mar;43(6):1015-1042. doi: 10.1038/s44318-024-00043-2. Epub 2024 Feb 15. EMBO J. 2024. PMID: 38360994 Free PMC article. - Monitoring Genomic Structural Rearrangements Resulting from Gene Editing.
Bailey SM, Cross EM, Kinner-Bibeau L, Sebesta HC, Bedford JS, Tompkins CJ. Bailey SM, et al. J Pers Med. 2024 Jan 19;14(1):110. doi: 10.3390/jpm14010110. J Pers Med. 2024. PMID: 38276232 Free PMC article. - Break-induced RNA-DNA hybrids (BIRDHs) in homologous recombination: friend or foe?
Gómez-González B, Aguilera A. Gómez-González B, et al. EMBO Rep. 2023 Dec 6;24(12):e57801. doi: 10.15252/embr.202357801. Epub 2023 Oct 11. EMBO Rep. 2023. PMID: 37818834 Free PMC article. Review. - Unrepaired base excision repair intermediates in template DNA strands trigger replication fork collapse and PARP inhibitor sensitivity.
Serrano-Benitez A, Wells SE, Drummond-Clarke L, Russo LC, Thomas JC, Leal GA, Farrow M, Edgerton JM, Balasubramanian S, Yang M, Frezza C, Gautam A, Brazina J, Burdova K, Hoch NC, Jackson SP, Caldecott KW. Serrano-Benitez A, et al. EMBO J. 2023 Sep 18;42(18):e113190. doi: 10.15252/embj.2022113190. Epub 2023 Jul 26. EMBO J. 2023. PMID: 37492888 Free PMC article.
References
- Branzei D, Foiani M (2005) The DNA damage response during DNA replication. Curr Opin Cell Biol 17: 568–575 - PubMed
- Gonzalez-Barrera S, Cortes-Ledesma F, Wellinger RE, Aguilera A (2003) Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol Cell 11: 1661–1671 - PubMed
- Ivanov D, Nasmyth K (2005) A topological interaction between cohesin rings and a circular minichromosome. Cell 122: 849–860 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous