Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions - PubMed (original) (raw)
Comparative Study
Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions
Luciana C Paulino et al. J Clin Microbiol. 2006 Aug.
Abstract
Psoriasis, a common cutaneous disease of unknown etiology, may be triggered by infections, including those due to fungi. Since the fungal community of human skin is poorly characterized, we aimed to analyze the mycological microbiota in healthy skin and psoriatic lesions. Twenty-five skin samples from five healthy subjects (flexor forearm) and three patients with psoriasis were analyzed using broad-range 18S ribosomal DNA (rDNA) and 5.8S rDNA/internal transcribed spacer 2 (ITS2) Malassezia-specific PCR primers. Broad-range PCR analysis indicated that most organisms resembled Malassezia. Malassezia-specific 5.8S/ITS2 analysis of 1,374 clones identified five species and four unknown phylotypes, potentially representing new species. The species distribution appears largely host specific and conserved in different sites of healthy skin. In three subjects, the Malassezia microbiota composition appeared relatively stable over time. Samples of Malassezia microbiota from healthy skin and psoriatic lesions were similar in one patient but substantially different in two others. These data indicate the predominance of Malassezia organisms in healthy human skin, host-specific variation, stability over time, and as yet, no consistent patterns differentiating psoriatic skin from healthy skin.
Figures
FIG. 1.
Fungal loci and sequences related to this study. (A) Schematic representation of the fungal ribosomal gene cluster, with PCR primers indicated (arrows). (B) Pairwise alignment between fragments of the 18S rRNA gene from human and representative fungal species, showing the sequences of primers EF4 and 1536 (long boxes). The human sequence contains a 7-bp insertion within the region represented by primer EF4 (small box). K, A/G; Y, T/C. (C) Pairwise alignment between fragments related to the 5.8S rRNA gene and ITS2 from humans and Malassezia species, showing the sequence of primers Mal1F and Mal1R. Bold letters in the human sequences refer to conserved nucleotides with the fungal sequences. H, T/C/A.
FIG. 2.
Neighbor-joining tree based on partial 18S rDNA sequences. The matrix of distances was calculated using the Jukes-Cantor algorithm. Bootstrap values are based on 500 replicates (values of at least 50% are shown). Organisms identified in this study are shown in bold. Codes correspond to GenBank accession numbers.
FIG. 3.
Neighbor-joining tree based on 5.8S rDNA and ITS2 sequences, showing the relationships among Malassezia organisms. The matrix of distances was calculated using the Jukes-Cantor algorithm. Bootstrap values are based on 500 replicates (values of at least 50% are shown). Organisms identified in this study are shown in bold. Codes correspond to GenBank accession numbers.
FIG. 4.
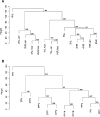
Consensus tree of hierarchical clustering of skin samples from healthy subjects (A) and patients with psoriasis (B). Height corresponds to Euclidian distance between samples. The number at each node represents the bootstrap value, based on 1,000 iterations. (A) 1N through 5N, healthy subjects; L, left arm; R, right arm. Samples L-Jan/R-Jan and L-Nov/R-Nov were obtained from the same site, 10 months apart. (B) 1P through 3P, patients with psoriasis; N, samples from normal skin; P1 through P3, samples from psoriatic lesions. Samples 1PP1A and 1PP1B were obtained from the same lesion, 6 months apart.
FIG. 5.
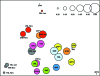
Scatterplot of the first two orthogonal principal axes based on the sample dissimilarity matrix. The samples from each subject are represented with the same color, and the sizes of the circles are proportional to the sample diversity, as determined by Rao's analysis. The diversity scale is shown in the upper right corner.
Similar articles
- Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR.
Paulino LC, Tseng CH, Blaser MJ. Paulino LC, et al. FEMS Yeast Res. 2008 May;8(3):460-71. doi: 10.1111/j.1567-1364.2008.00359.x. Epub 2008 Feb 20. FEMS Yeast Res. 2008. PMID: 18294199 - Molecular characterization of the skin fungal microbiome in patients with psoriasis.
Takemoto A, Cho O, Morohoshi Y, Sugita T, Muto M. Takemoto A, et al. J Dermatol. 2015 Feb;42(2):166-70. doi: 10.1111/1346-8138.12739. Epub 2014 Dec 15. J Dermatol. 2015. PMID: 25510344 - Molecular analysis of Malassezia microflora in the lesional skin of psoriasis patients.
Amaya M, Tajima M, Okubo Y, Sugita T, Nishikawa A, Tsuboi R. Amaya M, et al. J Dermatol. 2007 Sep;34(9):619-24. doi: 10.1111/j.1346-8138.2007.00343.x. J Dermatol. 2007. PMID: 17727364 - The role of Malassezia species in the ecology of human skin and as pathogens.
Guého E, Boekhout T, Ashbee HR, Guillot J, Van Belkum A, Faergemann J. Guého E, et al. Med Mycol. 1998;36 Suppl 1:220-9. Med Mycol. 1998. PMID: 9988511 Review. - Topographical and physiological differences of the skin mycobiome in health and disease.
Jo JH, Kennedy EA, Kong HH. Jo JH, et al. Virulence. 2017 Apr 3;8(3):324-333. doi: 10.1080/21505594.2016.1249093. Epub 2016 Oct 18. Virulence. 2017. PMID: 27754756 Free PMC article. Review.
Cited by
- Metagenomics: A new horizon in cancer research.
Banerjee J, Mishra N, Dhas Y. Banerjee J, et al. Meta Gene. 2015 Jun 14;5:84-9. doi: 10.1016/j.mgene.2015.05.005. eCollection 2015 Sep. Meta Gene. 2015. PMID: 26110115 Free PMC article. Review. - Antimicrobial active clothes display no adverse effects on the ecological balance of the healthy human skin microflora.
Hoefer D, Hammer TR. Hoefer D, et al. ISRN Dermatol. 2011;2011:369603. doi: 10.5402/2011/369603. Epub 2011 Apr 4. ISRN Dermatol. 2011. PMID: 22363849 Free PMC article. - Diverse Human Skin Fungal Communities in Children Converge in Adulthood.
Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI; NISC Comparative Sequencing Program; Segre JA, Kong HH. Jo JH, et al. J Invest Dermatol. 2016 Dec;136(12):2356-2363. doi: 10.1016/j.jid.2016.05.130. Epub 2016 Jul 29. J Invest Dermatol. 2016. PMID: 27476723 Free PMC article. - The human microbiome: our second genome.
Grice EA, Segre JA. Grice EA, et al. Annu Rev Genomics Hum Genet. 2012;13:151-70. doi: 10.1146/annurev-genom-090711-163814. Epub 2012 Jun 6. Annu Rev Genomics Hum Genet. 2012. PMID: 22703178 Free PMC article. Review. - Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples.
Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Lauber CL, et al. FEMS Microbiol Lett. 2010 Jun;307(1):80-6. doi: 10.1111/j.1574-6968.2010.01965.x. Epub 2010 Mar 25. FEMS Microbiol Lett. 2010. PMID: 20412303 Free PMC article.
References
- Baroni, A., I. Paoletti, E. Ruocco, M. Agozzino, M. A. Tufano, and G. Donnarumma. 2004. Possible role of Malassezia furfur in psoriasis: modulation of TGF-β1, integrin, and HSP70 expression in human keratinocytes and in the skin of psoriasis-affected patients. J. Cutan. Pathol. 31:35-42. - PubMed
- Baroni, A., B. Perfetto, I. Paoletti, E. Ruocco, N. Canozo, M. Orlando, and E. Buommino. 2001. Malassezia furfur invasiveness in a keratinocyte cell line (HaCat): effects on cytoskeleton and on adhesion molecule and cytokine expression. Arch. Dermatol. Res. 293:414-419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical