The outcomes of pathway database computations depend on pathway ontology - PubMed (original) (raw)
The outcomes of pathway database computations depend on pathway ontology
M L Green et al. Nucleic Acids Res. 2006.
Abstract
Different biological notions of pathways are used in different pathway databases. Those pathway ontologies significantly impact pathway computations. Computational users of pathway databases will obtain different results depending on the pathway ontology used by the databases they employ, and different pathway ontologies are preferable for different end uses. We explore differences in pathway ontologies by comparing the BioCyc and KEGG ontologies. The BioCyc ontology defines a pathway as a conserved, atomic module of the metabolic network of a single organism, i.e. often regulated as a unit, whose boundaries are defined at high-connectivity stable metabolites. KEGG pathways are on average 4.2 times larger than BioCyc pathways, and combine multiple biological processes from different organisms to produce a substrate-centered reaction mosaic. We compared KEGG and BioCyc pathways using genome context methods, which determine the functional relatedness of pairs of genes. For each method we employed, a pair of genes randomly selected from a BioCyc pathway is more likely to be related by that method than is a pair of genes randomly selected from a KEGG pathway, supporting the conclusion that the BioCyc pathway conceptualization is closer to a single conserved biological process than is that of KEGG.
Figures
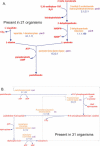
Figure 1
(A) Pathway for biosynthesis of pantothenate from 2-keto-isovalerate. (B) Pathway for biosynthesis of coenzyme A from pantothenate. Of the 204 BioCyc organisms, 31 include the pathway for biosynthesis of coenzyme A, but lack the pantothenate biosynthesis pathway. Another 21 organisms include the pathway for pantothenate biosynthesis, but lack the coenzyme A biosynthesis pathway. A total of 108 organisms include both pathways, while 44 lack both pathways.
Figure 2
Predicted pathway for teichoic acid biosynthesis in S.meliloti. Only where enzyme names and gene names are shown has an enzyme catalyzing a reaction in this pathway been identified in S.meliloti. Of the 204 BioCyc organisms, 155 include the branch of the teichoic acid biosynthesis pathway that synthesizes UDP-
d
-glucose. Only 38 organisms include any of the remaining reactions in the pathway, and only 31 of these include reactions from both branches.
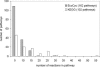
Figure 3
Size distribution of the KEGG and EcoCyc metabolic pathways.
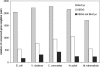
Figure 4
Number of conserved gene neighbors. Each pair of genes is selected randomly from a single KEGG metabolic map or from a single BioCyc metabolic pathway.
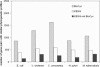
Figure 5
Number of gene pairs with similar phylogenetic profiles. Each pair of genes is selected randomly from a single KEGG metabolic map or from a single BioCyc metabolic pathway.
Figure 6
Number of gene pairs occurring in a predicted gene cluster. Each pair of genes is selected randomly from a single KEGG metabolic map or from a single BioCyc metabolic pathway.
Figure 7
Number of gene pairs related by gene fusion events. Each pair of genes is selected randomly from a single KEGG metabolic map or from a single BioCyc metabolic pathway.
Figure 8
Number of conserved gene neighbors and similar phylogenetic profiles randomly selected from EcoCyc superpathways compared to standard EcoCyc pathways and KEGG E.coli metabolic maps.
Similar articles
- Expansion of the BioCyc collection of pathway/genome databases to 160 genomes.
Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahrén D, Tsoka S, Darzentas N, Kunin V, López-Bigas N. Karp PD, et al. Nucleic Acids Res. 2005 Oct 24;33(19):6083-9. doi: 10.1093/nar/gki892. Print 2005. Nucleic Acids Res. 2005. PMID: 16246909 Free PMC article. - The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases.
Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. Caspi R, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D623-31. doi: 10.1093/nar/gkm900. Epub 2007 Oct 27. Nucleic Acids Res. 2008. PMID: 17965431 Free PMC article. - The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases.
Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD. Caspi R, et al. Nucleic Acids Res. 2010 Jan;38(Database issue):D473-9. doi: 10.1093/nar/gkp875. Epub 2009 Oct 22. Nucleic Acids Res. 2010. PMID: 19850718 Free PMC article. - Notes on the use of ontologies in the biochemical domain.
Rojas I, Ratsch E, Saric J, Wittig U. Rojas I, et al. In Silico Biol. 2004;4(1):89-96. Epub 2004 Mar 15. In Silico Biol. 2004. PMID: 15089756 Review. - Improvements to cardiovascular gene ontology.
Lovering RC, Dimmer EC, Talmud PJ. Lovering RC, et al. Atherosclerosis. 2009 Jul;205(1):9-14. doi: 10.1016/j.atherosclerosis.2008.10.014. Epub 2008 Nov 1. Atherosclerosis. 2009. PMID: 19046747 Free PMC article. Review.
Cited by
- Machine learning methods for metabolic pathway prediction.
Dale JM, Popescu L, Karp PD. Dale JM, et al. BMC Bioinformatics. 2010 Jan 8;11:15. doi: 10.1186/1471-2105-11-15. BMC Bioinformatics. 2010. PMID: 20064214 Free PMC article. - Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease.
Greenblum S, Turnbaugh PJ, Borenstein E. Greenblum S, et al. Proc Natl Acad Sci U S A. 2012 Jan 10;109(2):594-9. doi: 10.1073/pnas.1116053109. Epub 2011 Dec 19. Proc Natl Acad Sci U S A. 2012. PMID: 22184244 Free PMC article. - Critical assessment of human metabolic pathway databases: a stepping stone for future integration.
Stobbe MD, Houten SM, Jansen GA, van Kampen AH, Moerland PD. Stobbe MD, et al. BMC Syst Biol. 2011 Oct 14;5:165. doi: 10.1186/1752-0509-5-165. BMC Syst Biol. 2011. PMID: 21999653 Free PMC article. - The conservation and evolutionary modularity of metabolism.
Peregrín-Alvarez JM, Sanford C, Parkinson J. Peregrín-Alvarez JM, et al. Genome Biol. 2009;10(6):R63. doi: 10.1186/gb-2009-10-6-r63. Epub 2009 Jun 12. Genome Biol. 2009. PMID: 19523219 Free PMC article. - Genetics and epigenetics of rheumatoid arthritis.
Viatte S, Plant D, Raychaudhuri S. Viatte S, et al. Nat Rev Rheumatol. 2013 Mar;9(3):141-53. doi: 10.1038/nrrheum.2012.237. Epub 2013 Feb 5. Nat Rev Rheumatol. 2013. PMID: 23381558 Free PMC article. Review.
References
- Karp P.D., Paley S., Romero P. The pathway tools software. Bioinformatics. 2002;18:S225–S232. - PubMed
- Papin J.A., Stelling J., Price N.D., Klamt S., Schuster S., Palsson B.O. Comparison of network-based pathway analysis methods. Trends Biotechnol. 2004;22:400–405. - PubMed