Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length - PubMed (original) (raw)
Comparative Study
. 2006 Sep;16(9):1109-18.
doi: 10.1101/gr.5147406. Epub 2006 Aug 9.
Affiliations
- PMID: 16899654
- PMCID: PMC1557766
- DOI: 10.1101/gr.5147406
Comparative Study
Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length
Sergio Callejas et al. Genome Res. 2006 Sep.
Abstract
African trypanosomes are parasitic protozoa that infect a wide range of mammals, including humans. These parasites remain extracellular in the mammalian bloodstream, where antigenic variation allows them to survive the immune response. The Trypanosoma brucei nuclear genome sequence has been published recently. However, the significant chromosome size polymorphism observed among strains and subspecies of T. brucei, where total DNA content may vary up to 30%, necessitates a comparative study to determine the underlying basis and significance of such variation between parasites. In addition, the sequenced strain (Tb927) presents one of the smallest genomes analyzed among T. brucei isolates; therefore, establishing polymorphic regions will provide essential complementary information to the sequencing project. We have developed a Tb927 high-resolution DNA microarray to study DNA content variation along chromosome I, one of the most size-variable chromosomes, in different strains and subspecies of T. brucei. Results show considerable copy number polymorphism, especially at subtelomeres, but are insufficient to explain the observed size difference. Additional sequencing reveals that >50% of a larger chromosome I consists of arrays of variant surface glycoprotein genes (VSGs), involved in avoidance of acquired immunity. In total, the subtelomeres appear to be three times larger than the diploid core. These results reveal that trypanosomes can utilize subtelomeres for amplification and divergence of gene families to such a remarkable extent that they may constitute most of a chromosome, and that the VSG repertoire may be even larger than reported to date. Further experimentation is required to determine if these results are applicable to all size-variable chromosomes.
Figures
Figure 2.
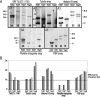
Validation of data derived from hybridization to the Tb927-_chr_Ia tiling path genomic microarray. (A) Hybridization of five gene-specific probes to Southern-blotted genomic fragments containing intact tandem arrays. Numbers at the bottom of each lane indicate (in kb) total DNA content of each array in each genome, estimated from these gels. Two bands in the same lane indicate that the array varies in length in the _chr_I homologs. The 10-kb band in panel 4 probably is due to cross-hybridization with a similar sequence in a different chromosome, and therefore has not been included in the final length for the _chr_I region containing pteridine transporter genes. DNA size markers are in kb. (B) Comparison between microarray results and Southern blot results. _y_-axis represents the amount of DNA compared with Tb927. Value of 1× in the _y_-axis indicates equal DNA content compared with Tb927.
Figure 1.
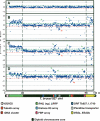
Copy number polymorphism in chromosome I homologs of T. brucei, assayed by comparative genomic hybridization to a microarray of T. brucei 927 _chr_Ia DNA fragments. (A) Co-hybridization of Tb927 genomic DNA differentially labeled with Cy5 and Cy3. (B) Cohybridization of differentially labeled Tb247 and Tb927 genomic DNA; (C) Tb427 and Tb927; (D) _T.b. gambiense_-M (Tbg-M) and Tb927. (Green and red lines) Two SD from the zero line in experiment A. Location of a fragment above the green line indicates copy number amplification in the test genome relative to Tb927; below the red line indicates DNA loss or sequence divergence in the test genome. (Vertical discontinuous lines) Subtelomere limits. Left subtelomere contains primarily retrotransposons (ingi) and gene families RHS and LRRP. Right subtelomere contains primarily genes involved in antigenic variation (_VSG_s) and associated genes (_ESAG_s). (_VSG_-ES) A VSG expression site; (FBP) a gene containing an F-box motif. (Green arrows) Gains of DNA; (red arrows) losses of DNA. Tandemly repeated gene coordinates: Tubulin array Tb927.1.2330–2410; Histone H3 array Tb927.1.2430–2550; Pteridine transporter array Tb927.1.2820, 2850, 2880; FBP array Tb927.1.4540–4650.
Figure 3.
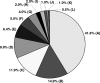
Identity of highest-scoring pairs for 192 sequence tags from clones selected from the Tb427-_chr_Ib-enriched plasmid library, derived by BLASTN comparison of sequence tags with Tb927 sequence in GeneDB and trypanosome sequence in the EMBL database. (A) _VSG_s, (B) VSG expression site-associated genes, (88% of these sequences are variants of ESAG3, most copies of which are associated with non-expressed VSG gene arrays in the Tb927 genome sequence [Berriman et al. 2005]). (C) intergenic sequences in VSG arrays in Tb927, (D) RHS genes and retrotransposons, (E) genes from other chromosomes, (F) UDP-dependent glycosyl transferase, (G) failed sequences, (H) _chr_I genes (chromosome core), (I) intergenic sequences (chromosome core), (J) telomeric and telomere-associated sequences, (K) no significant match, (L) UDP-gal-phosphoglycan transferase. (The genes in F and L are associated with VSG gene arrays in Tb927 [Berriman et al. 2005]. See Supplemental Table S2 for additional details.)
Figure 4.
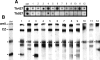
Hybridization of 12 Tb427_-chr_Ib-derived probes to dot-blots of Tb927 and Tb427 total DNA (A) and Southern blots of Tb427 chromosomes (B). In dot-blot experiments most probes hybridize only to Tb427, although in some cases the lower stringency of this experiment (compared with the initial screening) allows cross-hybridization to similar VSG families in Tb927. (Black dots) Hybridization to _chr_Ib (fully resolved in this gel), (black stars) hybridization to the _chr_Ia homolog (here compressed with other chromosomes), (_Chr_I) _chr_I-specific gene probe, (CZ) compression zone containing _chr_s VIIIb, IXb, Xab, and XIab (∼40% of megabase genome). See Supplemental Table S2 for additional information on each of the probes.
Figure 5.
Identity of highest-scoring pairs for sequence tags from 64 Tb427-_chr_VIa-specific (panel A) and 32 randomly selected clones from the Tb427-_chr_VIa-enriched plasmid library (panel B), derived by BLASTN comparison of sequence tags with Tb927 sequence in GeneDB and trypanosome sequence in the EMBL database. (A) _VSG_s, (B) VSG expression site-associated genes, (C) intergenic sequences in VSG arrays in Tb927, (D) RHS genes and retrotransposons, (E) genes from other chromosomes, (F) UDP-dependent glycosyl transferase, (G) _chr_VI genes (chromosome core), (H) intergenic sequences (chromosome core), (I) telomeric and telomere-associated sequences, (J) no significant match. See Supplemental Tables S3 and S4 for additional details.
Figure 6.
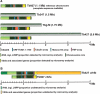
Models of chromosome expansion based on the comparative microarray analyses and sequencing of chromosome-specific DNA clones presented here. Note that we make the assumption that all gene families are positioned at the same location in both Tb927-_chr_Ia and Tb427-_chr_Ib, and therefore that arrays expand/contract in situ. (A) Comparison of c_hr_I size as estimated by PFGE analysis of different isolates of T. brucei (gray bars) with the theoretical maximum length estimated from the microarray and Southern blot analyses comparing the test chromosomes with Tb927-_chr_Ia. Colored regions correspond to tandemly repeated genes and gene families that show a significant degree of polymorphism compared with Tb927. (B) Our proposed model of Tb427-_chr_Ib organization. The central core is highly conserved compared with the sequenced Tb927-_chr_Ia; sequences in the left subtelomeric region are amplified about twofold, but there is no _VSG_-ES. Results presented here indicate that the remainder of the additional DNA in Tb427-_chr_Ib consists primarily of VSG arrays and associated sequences. We propose that this forms a hemizygous “subtelomeric” region of ∼2 Mb, and that this is probably on the right end of the chromosome.
Similar articles
- Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427.
Cross GA, Kim HS, Wickstead B. Cross GA, et al. Mol Biochem Parasitol. 2014 Jun;195(1):59-73. doi: 10.1016/j.molbiopara.2014.06.004. Epub 2014 Jun 30. Mol Biochem Parasitol. 2014. PMID: 24992042 - The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427.
Melville SE, Leech V, Navarro M, Cross GA. Melville SE, et al. Mol Biochem Parasitol. 2000 Dec;111(2):261-73. doi: 10.1016/s0166-6851(00)00316-9. Mol Biochem Parasitol. 2000. PMID: 11163435 - Variant Surface Glycoprotein gene repertoires in Trypanosoma brucei have diverged to become strain-specific.
Hutchinson OC, Picozzi K, Jones NG, Mott H, Sharma R, Welburn SC, Carrington M. Hutchinson OC, et al. BMC Genomics. 2007 Jul 13;8:234. doi: 10.1186/1471-2164-8-234. BMC Genomics. 2007. PMID: 17629915 Free PMC article. - The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes.
Horn D, Barry JD. Horn D, et al. Chromosome Res. 2005;13(5):525-33. doi: 10.1007/s10577-005-0991-8. Chromosome Res. 2005. PMID: 16132817 Review. - Mechanisms of antigenic variation in African trypanosomes.
Borst P, Rudenko G, Blundell PA, van Leeuwen F, Cross MA, McCulloch R, Gerrits H, Chaves IM. Borst P, et al. Behring Inst Mitt. 1997 Mar;(99):1-15. Behring Inst Mitt. 1997. PMID: 9303197 Review.
Cited by
- Genome plasticity driven by aneuploidy and loss of heterozygosity in Trypanosoma cruzi.
Cruz-Saavedra L, Schwabl P, Vallejo GA, Carranza JC, Muñoz M, Patino LH, Paniz-Mondolfi A, Llewellyn MS, Ramírez JD. Cruz-Saavedra L, et al. Microb Genom. 2022 Jun;8(6):mgen000843. doi: 10.1099/mgen.0.000843. Microb Genom. 2022. PMID: 35748878 Free PMC article. - Keeping Balance Between Genetic Stability and Plasticity at the Telomere and Subtelomere of Trypanosoma brucei.
Li B. Li B. Front Cell Dev Biol. 2021 Jul 5;9:699639. doi: 10.3389/fcell.2021.699639. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34291053 Free PMC article. Review. - TbSAP is a novel chromatin protein repressing metacyclic variant surface glycoprotein expression sites in bloodstream form Trypanosoma brucei.
Davies C, Ooi CP, Sioutas G, Hall BS, Sidhu H, Butter F, Alsford S, Wickstead B, Rudenko G. Davies C, et al. Nucleic Acids Res. 2021 Apr 6;49(6):3242-3262. doi: 10.1093/nar/gkab109. Nucleic Acids Res. 2021. PMID: 33660774 Free PMC article. - Read, Write, Adapt: Challenges and Opportunities during Kinetoplastid Genome Replication.
Damasceno JD, Marques CA, Black J, Briggs E, McCulloch R. Damasceno JD, et al. Trends Genet. 2021 Jan;37(1):21-34. doi: 10.1016/j.tig.2020.09.002. Epub 2020 Sep 28. Trends Genet. 2021. PMID: 32993968 Free PMC article. Review. - Genomic Organization and Generation of Genetic Variability in the RHS (Retrotransposon Hot Spot) Protein Multigene Family in Trypanosoma cruzi.
Bernardo WP, Souza RT, Costa-Martins AG, Ferreira ER, Mortara RA, Teixeira MMG, Ramirez JL, Da Silveira JF. Bernardo WP, et al. Genes (Basel). 2020 Sep 17;11(9):1085. doi: 10.3390/genes11091085. Genes (Basel). 2020. PMID: 32957642 Free PMC article.
References
- Barry J.D., McCulloch R., McCulloch R. Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. - PubMed
- Barry J.D., Ginger M.L., Burton P., McCulloch R., Ginger M.L., Burton P., McCulloch R., Burton P., McCulloch R., McCulloch R. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 2003;33:29–45. - PubMed
- Beals T.P., Boothroyd J.C., Boothroyd J.C. Genomic organization and context of a trypanosome variant surface glycoprotein gene family. J. Mol. Biol. 1992;225:961–971. - PubMed
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., Lennard N.J., Caler E., Hamlin N.E., Haas B., Caler E., Hamlin N.E., Haas B., Hamlin N.E., Haas B., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. . Science. 2005;309:416–422. - PubMed
- Biteau N., Bringaud F., Gibson W., Truc P., Baltz T., Bringaud F., Gibson W., Truc P., Baltz T., Gibson W., Truc P., Baltz T., Truc P., Baltz T., Baltz T. Characterization of Trypanozoon isolates using a repeated coding sequence and microsatellite markers. Mol. Biochem. Parasitol. 2000;105:185–201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources