Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors - PubMed (original) (raw)
Comparative Study
Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors
Gustavo A de Souza et al. Genome Biol. 2006.
Abstract
Background: The tear film is a thin layer of fluid that covers the ocular surface and is involved in lubrication and protection of the eye. Little is known about the protein composition of tear fluid but its deregulation is associated with disease states, such as diabetic dry eyes. This makes this body fluid an interesting candidate for in-depth proteomic analysis.
Results: In this study, we employ state-of-the-art mass spectrometric identification, using both a hybrid linear ion trap-Fourier transform (LTQ-FT) and a linear ion trap-Orbitrap (LTQ-Orbitrap) mass spectrometer, and high confidence identification by two consecutive stages of peptide fragmentation (MS/MS/MS or MS3), to characterize the protein content of the tear fluid. Low microliter amounts of tear fluid samples were either pre-fractionated with one-dimensional SDS-PAGE and digested in situ with trypsin, or digested in solution. Five times more proteins were detected after gel electrophoresis compared to in solution digestion (320 versus 63 proteins). Ontology classification revealed that 64 of the identified proteins are proteases or protease inhibitors. Of these, only 24 have previously been described as components of the tear fluid. We also identified 18 anti-oxidant enzymes, which protect the eye from harmful consequences of its exposure to oxygen. Only two proteins with this activity have been previously described in the literature.
Conclusion: Interplay between proteases and protease inhibitors, and between oxidative reactions, is an important feature of the ocular environment. Identification of a large set of proteins participating in these reactions may allow discovery of molecular markers of disease conditions of the eye.
Figures
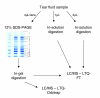
Figure 1
Approach used for tear fluid analysis. The tear fluid was analyzed by both in-solution digestion (1 and 4 μl) and one-dimensional gel separation combined with MS (GeLC-MS; 2 lanes of 4 μl each) on a LTQ-FT, and also through GeLC-MS on a LTQ-Orbitrap. The numbers indicate the bands according to the slicing pattern used for sample fractionation prior to in situ digestion.
Figure 2
LTQ-FT data for the peptide at m/z 494.29 (ILDLIESGK). The figure shows an example of data-dependent acquisition on the LTQ-FT. (a) SIM scan of the doubly charged peptide at 494.29, observed in the total ion chromatogram. (b) The peptide is selected for fragmentation and MS2 acquisition, and (c) the most intense daughter-ion is selected for a new round of fragmentation MS3. Partial data obtained in the MS3 is used to confirm sequence observed in the MS2 and, consequently, improves the probability score for the identified sequence.
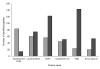
Figure 3
Data comparison between LTQ-FT and LTQ-Orbitrap spectrometry. The numbers of peptides for the top six identified proteins (LTQ-FT data) were compared between the two methods. Except for the protein Apolipoprotein B100, we observed a significant increase in the number of peptides identified with the LTQ-Orbitrap. This pattern was observed for most of the proteins identified with more then three peptides in the LTQ-FT. Light gray bars represent LTQ-FT data, dark gray represents LTQ-Orbitrap data.
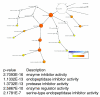
Figure 4
Statistical analysis of GO classification using the BiNGO tool. After identification and merging of the two datasets by the Protein Center tool, a gene list of the 491 identified proteins was generated and submitted to the BiNGO tool. This tool is able to apply statistical analysis to determine over-represented groups present in the sample. The figure shows a partial diagram of the analysis of GO molecular function, zoomed in the protease inhibitors branch. The p values for this group are also indicated.
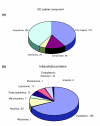
Figure 5
GO classification of tear fluid based on cellular localization. (a) Of the 491 proteins identified in the tear fluid, 200 were classified as intracellular proteins, while only 68 were classified as extracellular. As already described in the literature, the presence of intracellular proteins may be a result of cell death in the epithelium in close contact with the eye. (b) From the intracellular group, the great majority of proteins belongs to the cytoplasmic region, with some organelles being well represented, such as the lysosome (BiNGO p value of 6.9216E-8, the third highest score after cytoplasmatic and extracellular proteins).
Figure 6
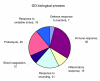
Relevant GO biological processes identified in the tear fluid. In the tear fluid, the most over-represented groups identified according to GO biological process were those involved in defense of the eye environment. These mainly comprised process such as immune response, defense against external biotic agents, response to wounding, and blood coagulation. Interestingly, 18 proteins responsible for response to oxidative stress were identified, only two of them described previously in tear fluid samples.
Similar articles
- The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins.
Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. Adachi J, et al. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-R80. Genome Biol. 2006. PMID: 16948836 Free PMC article. - Comparison of Different Mass Spectrometry Workflows for the Proteomic Analysis of Tear Fluid.
Jones G, Lee TJ, Glass J, Rountree G, Ulrich L, Estes A, Sezer M, Zhi W, Sharma S, Sharma A. Jones G, et al. Int J Mol Sci. 2022 Feb 19;23(4):2307. doi: 10.3390/ijms23042307. Int J Mol Sci. 2022. PMID: 35216421 Free PMC article. - Investigation of the human tear film proteome using multiple proteomic approaches.
Green-Church KB, Nichols KK, Kleinholz NM, Zhang L, Nichols JJ. Green-Church KB, et al. Mol Vis. 2008 Mar 7;14:456-70. Mol Vis. 2008. PMID: 18334958 Free PMC article. - Critical role of mass spectrometry proteomics in tear biomarker discovery for multifactorial ocular diseases (Review).
Ma JYW, Sze YH, Bian JF, Lam TC. Ma JYW, et al. Int J Mol Med. 2021 May;47(5):83. doi: 10.3892/ijmm.2021.4916. Epub 2021 Mar 24. Int J Mol Med. 2021. PMID: 33760148 Free PMC article. Review. - Tear analysis in ocular surface diseases.
Zhou L, Beuerman RW. Zhou L, et al. Prog Retin Eye Res. 2012 Nov;31(6):527-50. doi: 10.1016/j.preteyeres.2012.06.002. Epub 2012 Jun 23. Prog Retin Eye Res. 2012. PMID: 22732126 Review.
Cited by
- An excitation wavelength-optimized, stable SERS biosensing nanoplatform for analyzing adenoviral and AstraZeneca COVID-19 vaccination efficacy status using tear samples of vaccinated individuals.
Kim W, Kim S, Han J, Kim TG, Bang A, Choi HW, Min GE, Shin JH, Moon SW, Choi S. Kim W, et al. Biosens Bioelectron. 2022 May 15;204:114079. doi: 10.1016/j.bios.2022.114079. Epub 2022 Feb 8. Biosens Bioelectron. 2022. PMID: 35151942 Free PMC article. - Comparative analysis of the tear protein profile in mycotic keratitis patients.
Ananthi S, Chitra T, Bini R, Prajna NV, Lalitha P, Dharmalingam K. Ananthi S, et al. Mol Vis. 2008 Mar 12;14:500-7. Mol Vis. 2008. PMID: 18385783 Free PMC article. - Maspin increases extracellular plasminogen activator activity associated with corneal fibroblasts and myofibroblasts.
Warejcka DJ, Narayan M, Twining SS. Warejcka DJ, et al. Exp Eye Res. 2011 Nov;93(5):618-27. doi: 10.1016/j.exer.2011.07.008. Epub 2011 Jul 27. Exp Eye Res. 2011. PMID: 21810423 Free PMC article. - Comparative analysis of human tear fluid and aqueous humor proteomes.
Beisel A, Jones G, Glass J, Lee TJ, Töteberg-Harms M, Estes A, Ulrich L, Bollinger K, Sharma S, Sharma A. Beisel A, et al. Ocul Surf. 2024 Jul;33:16-22. doi: 10.1016/j.jtos.2024.03.011. Epub 2024 Mar 30. Ocul Surf. 2024. PMID: 38561100 - Biomolecular Evidence of Silk from 8,500 Years Ago.
Gong Y, Li L, Gong D, Yin H, Zhang J. Gong Y, et al. PLoS One. 2016 Dec 12;11(12):e0168042. doi: 10.1371/journal.pone.0168042. eCollection 2016. PLoS One. 2016. PMID: 27941996 Free PMC article.
References
- Dilly PN. Structure and function of the tear film. Adv Exp Med Biol. 1994;350:239–247. - PubMed
- Fleiszig SM, McNamara NA, Evans DJ. The tear film and defense against infection. Adv Exp Med Biol. 2002;506:523–530. - PubMed
- Kijlstra A, Kuizenga A. Analysis and function of the human tear proteins. Adv Exp Med Biol. 1994;350:299–308. - PubMed
- Grus FH, Augustin AJ. High performance liquid chromatography analysis of tear protein patterns in diabetic and non-diabetic dry-eye patients. Eur J Ophthalmol. 2001;11:19–24. - PubMed
- Herber S, Grus FH, Sabuncuo P, Augustin AJ. Changes in the tear protein patterns of diabetic patients using two-dimensional electrophoresis. Adv Exp Med Biol. 2002;506:623–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical