The structure and function of frataxin - PubMed (original) (raw)
Review
The structure and function of frataxin
Krisztina Z Bencze et al. Crit Rev Biochem Mol Biol. 2006 Sep-Oct.
Abstract
Frataxin, a highly conserved protein found in prokaryotes and eukaryotes, is required for efficient regulation of cellular iron homeostasis. Humans with a frataxin deficiency have the cardio- and neurodegenerative disorder Friedreich's ataxia, commonly resulting from a GAA trinucleotide repeat expansion in the frataxin gene. While frataxin's specific function remains a point of controversy, the general consensus is that the protein assists in controlling cellular iron homeostasis by directly binding iron. This review focuses on the structural and biochemical aspects of iron binding by the frataxin orthologs and outlines molecular attributes that may help explain the protein's role in different cellular pathways.
Figures
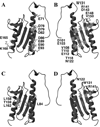
Figure 1
Top: ribbon diagram for yeast, human and bacterial frataxin. Middle: electropotential plots for proteins in same orientation. Bottom: electropotential plots for proteins rotated −90 degrees around the y-axis compared to top display. Structure figures made using solution structures of Yfh1 (PDB ID# 2GA5), HsFtx (PDB ID# 1LY7) and CyaY (PDB ID# 1SOY) frataxins.
Figure 2
ClustalX alignment for a subset of characterized frataxin orthologs. Bottom three sequences represent structurally characterized frataxin orthologs. Secondary structural elements and ruler representing Yfh1 properties are given below the sequences.
Figure 3
Yfh1 residues that are highly conserved on the helical (A) and β-sheet (B) planes of the protein. Identity of HsFtx FRDA point mutations on the helical (C) and β-sheet (D) planes of Yfh1. Structure figures made using Yfh1 solution structure (PDB ID# 2GA5).
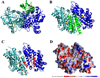
Figure 4
Lowest energy simulation of Yfh1 monomer docked to the metal loaded yeast ferrochelatase. (A)Side view of single Yfh1 monomer (green) docked to yeast ferrochelatase dimer (dark and light blue). Co2+ is bound in the yeast ferrochelatase structure in the assembly active site close to the four membrane attachment lips at the bottom of the figure. (B) Side view (90°—Horizontal rotation of Figure 4A) showing monomeric Yfh1 interacts with both units in the ferrochelatase dimer. (C) Ferrochelatase side view with Yfh1 structure removed to show the residues that directly interact with frataxin (in red). (D) Electrostatic potential plots (calculated and vendered with Grasp) of ferrochelatase, side view. Figures A, B, and C prepared using VMD (Humphry et al., 1996). Docking simulations were performed using ZDock (Chen et al., 2003) using the Yfh1 structure simulations (PDB ID# 2GA5) and Co2+ loaded yeast ferrochelatase structure (PDB ID# 1L8X).
Similar articles
- Frataxin, a molecule of mystery: trading stability for function in its iron-binding site.
Lane DJ, Richardson DR. Lane DJ, et al. Biochem J. 2010 Feb 9;426(2):e1-3. doi: 10.1042/BJ20091959. Biochem J. 2010. PMID: 20141512 - A structural approach to understanding the iron-binding properties of phylogenetically different frataxins.
Adinolfi S, Trifuoggi M, Politou AS, Martin S, Pastore A. Adinolfi S, et al. Hum Mol Genet. 2002 Aug 1;11(16):1865-77. doi: 10.1093/hmg/11.16.1865. Hum Mol Genet. 2002. PMID: 12140189 - Iron-binding activity in yeast frataxin entails a trade off with stability in the alpha1/beta1 acidic ridge region.
Correia AR, Wang T, Craig EA, Gomes CM. Correia AR, et al. Biochem J. 2010 Feb 9;426(2):197-203. doi: 10.1042/BJ20091612. Biochem J. 2010. PMID: 20001966 Free PMC article. - Exploring frataxin function.
Busi MV, Gomez-Casati DF. Busi MV, et al. IUBMB Life. 2012 Jan;64(1):56-63. doi: 10.1002/iub.577. Epub 2011 Nov 17. IUBMB Life. 2012. PMID: 22095894 Review. - Friedreich's ataxia.
Alper G, Narayanan V. Alper G, et al. Pediatr Neurol. 2003 May;28(5):335-41. doi: 10.1016/s0887-8994(03)00004-3. Pediatr Neurol. 2003. PMID: 12878293 Review.
Cited by
- Probing the kinetic stabilities of Friedreich's ataxia clinical variants using a solid phase GroEL chaperonin capture platform.
Correia AR, Naik S, Fisher MT, Gomes CM. Correia AR, et al. Biomolecules. 2014 Oct 20;4(4):956-79. doi: 10.3390/biom4040956. Biomolecules. 2014. PMID: 25333765 Free PMC article. - Friedreich's ataxia: pathology, pathogenesis, and molecular genetics.
Koeppen AH. Koeppen AH. J Neurol Sci. 2011 Apr 15;303(1-2):1-12. doi: 10.1016/j.jns.2011.01.010. J Neurol Sci. 2011. PMID: 21315377 Free PMC article. Review. - Extra-mitochondrial mouse frataxin and its implications for mouse models of Friedreich's ataxia.
Weng L, Laboureur L, Wang Q, Guo L, Xu P, Gottlieb L, Lynch DR, Mesaros C, Blair IA. Weng L, et al. Sci Rep. 2020 Sep 25;10(1):15788. doi: 10.1038/s41598-020-72884-w. Sci Rep. 2020. PMID: 32978498 Free PMC article. - Frataxin and mitochondrial FeS cluster biogenesis.
Stemmler TL, Lesuisse E, Pain D, Dancis A. Stemmler TL, et al. J Biol Chem. 2010 Aug 27;285(35):26737-26743. doi: 10.1074/jbc.R110.118679. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522547 Free PMC article. Review. - Defects in mitochondrial axonal transport and membrane potential without increased reactive oxygen species production in a Drosophila model of Friedreich ataxia.
Shidara Y, Hollenbeck PJ. Shidara Y, et al. J Neurosci. 2010 Aug 25;30(34):11369-78. doi: 10.1523/JNEUROSCI.0529-10.2010. J Neurosci. 2010. PMID: 20739558 Free PMC article.
References
- Adinolfi S, Trifuoggi M, Politou AS, Martin S, Pastore A. A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Hum. Mol. Genet. 2002;11:1865. - PubMed
- Adinolfi S, Nair M, Politou A, Bayer E, Martin S, Temussi P, Pastore A. The factors governing the thermal stability of frataxin orthologues: how to increase a protein’s stability. Biochemistry. 2004;43:6511. - PubMed
- Adinolfi S, Rizzo F, Masino L, Nair M, Martin SR, Pastore A, Temussi PA. Bacterial IscU is a well folded and functional single domain protein. Eur J Biochem. 2004;271:2093. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous