Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis - PubMed (original) (raw)
Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis
Dongfang Wang et al. Proc Natl Acad Sci U S A. 2006.
Abstract
In Arabidopsis, a complex of Polycomb-group (PcG) proteins functions in the female gametophyte to control the initiation of seed development. Mutations in the PcG genes, including MEDEA (MEA) and FERTILIZATION-INDEPENDENT SEED 2 (FIS2), produce autonomous seeds where endosperm proliferation occurs in the absence of fertilization. By using a yeast two-hybrid screen, we identified MEA and a related protein, SWINGER (SWN), as SET-domain partners of FIS2. Localization data indicated that all three proteins are present in the female gametophyte. Although single-mutant swn plants did not show any defects, swn mutations enhanced the mea mutant phenotype in producing autonomous seeds. Thus, MEA and SWN perform partially redundant functions in controlling the initiation of endosperm development before fertilization in Arabidopsis.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
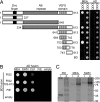
Interaction of FIS2 with SET-domain PcG proteins in yeast and in vitro. (A) Mapping MEA-interacting domains of FIS2 by using Y2H assays. The deletion constructs of FIS2 are shown schematically with numbers indicating the position of amino acids included in each construct. Diploid yeast harboring both pGBK (BD) and pGAD (AD) constructs were spotted on QDO (synthetic complete medium minus leucine, tryptophan, adenine, and histidine) and DDO (synthetic complete medium minus leucine and tryptophan). (B) Interaction of FIS2 and FIE with three SET-domain PcG proteins using Y2H assays is indicated by the growth of yeast on QDO medium. empty, empty pGBK, or pGAD vector. (C) In vitro binding of myc-tagged FIS2 (641–813) polypeptide with HA-tagged full-length MEA, SWN, and FIE labeled with [35S]methionine. The input lane (i) corresponds to 5% of the protein used for in vitro coimmunoprecipitation assays in output lanes (o).
Fig. 2.
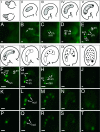
Expression of SWN-GFP, FIS2-YFP, and MEA-YFP during FG and early seed development. (I–X) Schematic representations of stages of FG and early endosperm development in Arabidopsis. The FG is formed from a surviving megaspore after the megaspore mother cell undergoes meiosis (I). The shaded areas in II–VI represent the developing FGs (26). Dividing endosperm nuclei during early seed development are depicted in VII–X. (A–T) Epifluorescence images of GFP/YFP activity corresponding to stages shown in I–V (A–E) and VI–X (F–J, K–O, and P–T). Images were obtained with the focal planes through the megaspore mother cell (A), developing FG (B–F, K, and P), or developing endosperm (G–J, L–O, and Q–T). (A–J) SWN-GFP expression. In most transgenic plants, SWN-GFP expression was not detectable in the endosperm (G–H). (K–O) FIS2-YFP expression. (P–T) MEA-YFP expression. an, antipodal cell nucleus; cn, chalazal nucleus; ccn, central cell nucleus; dsn, degenerating synergid nucleus; ecn, egg cell nucleus; en, endosperm nucleus; fmn, functional megaspore nucleus; mn, micropylar nucleus; ncd, nuclear cytoplasmic domain; pn, polar nucleus; sn, synergid cell nucleus; zn, zygote nucleus. [Scale bars: 10 μm (A–E); 30 μm (F–T).]
Fig. 3.
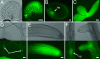
Expression of SWN-GFP in ovule, seed, and vegetative structures. (A) Bright-field (Left) and the corresponding epifluorescence (Right) images of a flower stage 13 (44) ovule showing SWN-GFP in all nuclei of the integument cells. The focal plane is through the outer integument. (B) Epifluorescence image of a developing seed containing a heart-stage embryo. (C) Epifluorescence image of a bent cotyledon-stage embryo dissected away from the endosperm and seed coat. (D) Bright-field (Upper) and the corresponding epifluorescence (Lower) images of a trichome on the surface of a rosette leaf. (E) Bright-field (Upper) and the corresponding epifluorescence (Lower) images of a root tip. (F) Bright-field (Upper) and the corresponding epifluorescence (Lower) images of a lateral root primordium and a lateral root. he, heart stage embryo; lrp, lateral root primordium; tn, trichome cell nucleus. (Scale bars: 30 μm.)
Similar articles
- The Polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis.
Schmidt A, Wöhrmann HJ, Raissig MT, Arand J, Gheyselinck J, Gagliardini V, Heichinger C, Walter J, Grossniklaus U. Schmidt A, et al. Plant J. 2013 Mar;73(5):776-87. doi: 10.1111/tpj.12070. Epub 2013 Feb 12. Plant J. 2013. PMID: 23146178 - Sexual and apomictic seed formation in Hieracium requires the plant polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM.
Rodrigues JC, Tucker MR, Johnson SD, Hrmova M, Koltunow AM. Rodrigues JC, et al. Plant Cell. 2008 Sep;20(9):2372-86. doi: 10.1105/tpc.108.059287. Epub 2008 Sep 23. Plant Cell. 2008. PMID: 18812497 Free PMC article. - The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1.
Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. Köhler C, et al. Genes Dev. 2003 Jun 15;17(12):1540-53. doi: 10.1101/gad.257403. Genes Dev. 2003. PMID: 12815071 Free PMC article. - Seed development: with or without sex?
Ma H. Ma H. Curr Biol. 1999 Sep 9;9(17):R636-9. doi: 10.1016/s0960-9822(99)80411-4. Curr Biol. 1999. PMID: 10508576 Review. - [Genomic imprinting and seed development].
Zhang WW, Cao SX, Jiang L, Zhu SS, Wan JM. Zhang WW, et al. Yi Chuan. 2005 Jul;27(4):665-70. Yi Chuan. 2005. PMID: 16120596 Review. Chinese.
Cited by
- H3K36 methyltransferase GhKMT3;1a and GhKMT3;2a promote flowering in upland cotton.
Ju J, Li Y, Ling P, Luo J, Wei W, Yuan W, Wang C, Su J. Ju J, et al. BMC Plant Biol. 2024 Aug 2;24(1):739. doi: 10.1186/s12870-024-05457-y. BMC Plant Biol. 2024. PMID: 39095699 Free PMC article. - Comprehensive Survey of ChIP-Seq Datasets to Identify Candidate Iron Homeostasis Genes Regulated by Chromatin Modifications.
Yu Y, Wang Y, Yao Z, Wang Z, Xia Z, Lee J. Yu Y, et al. Methods Mol Biol. 2023;2665:95-111. doi: 10.1007/978-1-0716-3183-6_9. Methods Mol Biol. 2023. PMID: 37166596 - Genome-wide identification and characterization of polycomb repressive complex 2 core components in upland cotton (Gossypium hirsutum L.).
Cheng K, Lei C, Zhang S, Zheng Q, Wei C, Huang W, Xing M, Zhang J, Zhang X, Zhang X. Cheng K, et al. BMC Plant Biol. 2023 Feb 1;23(1):66. doi: 10.1186/s12870-023-04075-4. BMC Plant Biol. 2023. PMID: 36721081 Free PMC article. - Histone Modification and Chromatin Remodeling During the Seed Life Cycle.
Ding X, Jia X, Xiang Y, Jiang W. Ding X, et al. Front Plant Sci. 2022 Apr 25;13:865361. doi: 10.3389/fpls.2022.865361. eCollection 2022. Front Plant Sci. 2022. PMID: 35548305 Free PMC article. Review. - Genome-Wide Identification and Characterization of SET Domain Family Genes in Brassica napus L.
Sehrish S, Sumbal W, Xie M, Zhao C, Zuo R, Gao F, Liu S. Sehrish S, et al. Int J Mol Sci. 2022 Feb 9;23(4):1936. doi: 10.3390/ijms23041936. Int J Mol Sci. 2022. PMID: 35216050 Free PMC article.
References
- Costa L. M., Gutierrez-Marcos J. F., Dickinson H. G. Trends Plant Sci. 2004;9:507–514. - PubMed
- Grossniklaus U., Vielle-Calzada J. P., Hoeppner M. A., Gagliano W. B. Science. 1998;280:446–450. - PubMed
- Guitton A. E., Page D. R., Chambrier P., Lionnet C., Faure J. E., Grossniklaus U., Berger F. Development (Cambridge, U.K.) 2004;131:2971–2981. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases