Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease - PubMed (original) (raw)
Case Reports
doi: 10.1371/journal.pgen.0020131.
Stéphanie Boisson-Dupuis, Emmanuelle Jouanguy, Guillaume Vogt, Jacqueline Feinberg, Ada Prochnicka-Chalufour, Armanda Casrouge, Kun Yang, Claire Soudais, Claire Fieschi, Orchidée Filipe Santos, Jacinta Bustamante, Capucine Picard, Ludovic de Beaucoudrey, Jean-François Emile, Peter D Arkwright, Robert D Schreiber, Claudia Rolinck-Werninghaus, Angela Rösen-Wolff, Klaus Magdorf, Joachim Roesler, Jean-Laurent Casanova
Affiliations
- PMID: 16934001
- PMCID: PMC1550284
- DOI: 10.1371/journal.pgen.0020131
Case Reports
Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease
Ariane Chapgier et al. PLoS Genet. 2006.
Abstract
The transcription factor signal transducer and activator of transcription-1 (STAT1) plays a key role in immunity against mycobacterial and viral infections. Here, we characterize three human STAT1 germline alleles from otherwise healthy patients with mycobacterial disease. The previously reported L706S, like the novel Q463H and E320Q alleles, are intrinsically deleterious for both interferon gamma (IFNG)-induced gamma-activating factor-mediated immunity and interferon alpha (IFNA)-induced interferon-stimulated genes factor 3-mediated immunity, as shown in STAT1-deficient cells transfected with the corresponding alleles. Their phenotypic effects are however mediated by different molecular mechanisms, L706S affecting STAT1 phosphorylation and Q463H and E320Q affecting STAT1 DNA-binding activity. Heterozygous patients display specifically impaired IFNG-induced gamma-activating factor-mediated immunity, resulting in susceptibility to mycobacteria. Indeed, IFNA-induced interferon-stimulated genes factor 3-mediated immunity is not affected, and these patients are not particularly susceptible to viral disease, unlike patients homozygous for other, equally deleterious STAT1 mutations recessive for both phenotypes. The three STAT1 alleles are therefore dominant for IFNG-mediated antimycobacterial immunity but recessive for IFNA-mediated antiviral immunity at the cellular and clinical levels. These STAT1 alleles define two forms of dominant STAT1 deficiency, depending on whether the mutations impair STAT1 phosphorylation or DNA binding.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. Pathways of IFNG-Induced GAF-Mediated Immunity and IFNA-Induced ISGF3-Mediated Immunity
Patients homozygous for null STAT1 mutations [36,37] suffer from both mycobacterial and viral diseases. Patients heterozygous for STAT1 mutations L706S, Q463H, and E320Q suffer from mycobacterial but not viral diseases ([17] and this report).
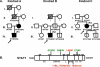
Figure 2. Novel STAT1 Mutations in Two Kindreds
(A) STAT1 genotype and clinical phenotype of three kindreds. In kindred A, members I.1 and II.1 had tuberculosis, and III.2 and IV.1 (P1) had severe BCG disease. In kindred B, members I.1 and III. 2 (P2) were infected with M. tuberculosis and M. avium, respectively. Kindred C has been described elsewhere; II.2 (P3) developed disseminated BCG disease. Individuals with clinical disease caused by weakly virulent (BCG or M. avium) and more virulent (M. tuberculosis) mycobacteria are indicated in black and gray, respectively, and healthy individuals are shown in white. The index cases are indicated with an arrow. Genetically affected individuals (heterozygous for any of the three STAT1 mutations) with no clinical phenotype at the time of this study are indicated by a vertical line. Known STAT1 genotypes (WT, E320Q, Q463H, L706S) are indicated under each individual, with a question mark indicating unknown genotype. (B) The human STAT1 coding region is shown, with its known pathogenic mutations. The coiled-coil domain (CC), DNA-binding domain (DNA-B), linker domain (L), SH2 domain (SH2), tail segment domain (TS), and _trans_-activator domain (TA) are indicated, together with their amino-acid boundaries. Tyrosine 701 (Y) is also indicated. Mutations in red are recessive mutations associated with complete STAT1 deficiency (due to a lack of STAT1 production), impaired IFNG-induced GAF activation and IFNA-induced ISGF3 activation, and a syndrome of predisposition to mycobacterial and severe viral disease in homozygous individuals. Mutations in green are associated, in heterozygous individuals, with partial STAT1 deficiency (normal STAT1 expression), impaired IFNG-induced GAS-binding activity but normal IFNA-induced ISRE-binding activity, and MSMD (predisposition to mycobacterial but not viral disease). Mutations reported for the first time in this study are indicated in italics.
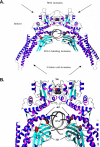
Figure 3. Molecular Representation of STAT1 Mutants
(A) Ribbon representation of the WT STAT1 homodimer complexed with DNA. Secondary structure elements representing the β strands are shown in cyan, and the helices are shown in blue-magenta. Atoms of residues in mutated positions L706, E320, and Q463 (indicated by arrows) are shown in space-filling models. Atoms are shown in red (for oxygen), blue (for nitrogen), and gray (for carbon). (B) Magnified focus on the region containing the three mutated residues.
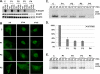
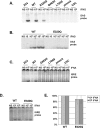
Figure 4. Normal Activation but Impaired DNA-Binding Activity of STAT1 in Heterozygous Cells from Patients
(A) Western blot of total protein extracts (100 μg) from EBV-transformed B cells derived from a healthy control (C), three patients under study (P1, P2, P3), and a patient with recessive complete STAT1 deficiency (P4) homozygous for the 1758_1759delAG mutation, probed with specific antibodies against phosphorylated-Tyr-701-STAT1, STAT1, and STAT3. EBV-transformed B cells were not stimulated (NS) or were stimulated with IFNA or IFNG (105 IU/ml) for 30 min. (B) Immunofluorescence staining with a STAT1-specific antibody of skin-derived SV40-transformed fibroblasts from a healthy control (C) and three patients under study (P1, P2, P3). Fibroblasts were not stimulated (NS) or were stimulated with IFNA or IFNG (105 IU/ml) for 30 min. (C and E) EMSA of nuclear extracts (5 μg) from EBV-transformed B cells derived from a healthy control (C), three patients under study (P1, P2, P3), and the patient with complete STAT1 deficiency (P4). EBV-transformed B cells were not stimulated (NS) or were stimulated for 30 min with 103 and 105 IU/ml of IFNG (C) and IFNA (E), respectively. Radiolabeled GAS (C) or ISRE (E) probes were used. (D) Quantification of four to six independent experiments by PhosphoImager SI (Molecular Dynamics, Piscataway, New Jersey, United States) using the GAS probe in response to 105 IU/ml of IFNG is also presented. The mean, minimum, and maximum values are expressed with respect to the positive control response (100%). For (A–C) and (E), one experiment representative of three to five independent experiments is shown.
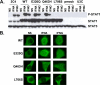
Figure 5. Normal Activation of STAT1 Mutants in Stable Transfectants
(A) Western blot of total protein extracts (100 μg) from a parental fibrosarcoma cell line (2C4) and STAT1-deficient U3C fibrosarcoma cell clones, untransfected (U3C) or stably cotransfected with a zeocin-resistance vector and a vector containing a mock (pmock), WT, E320Q, Q463H, or L706S STAT1 allele, with antibodies specific for phosphorylated-Tyr-701-STAT1, STAT1, and STAT3. The cells were not stimulated (NS) or were stimulated for 30 min with 105 IU/ml IFNA or IFNG. (B) Immunofluorescence staining, with a STAT1-specific antibody, of STAT1-deficient U3C fibrosarcoma cell clones, stably cotransfected with a zeocin-resistance vector and a vector containing a WT, E320Q, Q463H, or L706S STAT1 allele. The cells were not stimulated (NS) or were stimulated with IFNA or IFNG (105 IU/ml) for 30 min. For (A) and (B), one experiment representative of three independent experiments is shown.
Figure 6. Impaired DNA-Binding Activity of STAT1 Mutants in Stable Transfectants
(A–D) EMSA of nuclear extracts (5 μg [A and B]; 30 μg [C and D]) from a parental fibrosarcoma cell line (2C4) and STAT1-deficient U3C fibrosarcoma cell clones, untransfected (U3C) or stably cotransfected with a zeocin-resistance vector and a vector containing a mock (pmock), WT, E320Q, Q463H, or L706S STAT1 allele. The cells were not stimulated (NS) or were stimulated for 30 min with the indicated doses of IFNG (A and B) or IFNA (C and D). We used the radiolabeled GAS probe FCGR1 (A and B) or an ISRE probe (C and D). For (A–D), one experiment representative of three to five independent experiments is shown. (E) Quantification of three independent experiments by PhosphoImager SI (Molecular Dynamics), using the ISRE probe, in response to 104 and 105 IU/ml IFNA is also presented. The mean, minimum, and maximum values are expressed with respect to the WT stable transfectant clone response (100%).
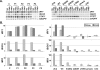
Figure 7. Impact of STAT1 Mutations on Transcription
(A) Levels of mRNA corresponding to STAT1, IFNG- and/or IFNA-inducible genes (IRF1 and ISG15) and GADP in EBV-transformed B cells from a control (C), the three affected individuals (P1, P2, and P3) and a STAT1-deficient individual (P4), or in the 2C4 parental fibrosarcoma cell line, STAT1-deficient U3C cell line, and U3C cells stably transfected with a mock (pmock), WT, E320Q, Q463H, or L706S STAT1 allele, either not stimulated (NS), or stimulated for 2 h with 103 of IFNG or 104 of IFNA for EBV-transformed B cells and 105 IU/ml of IFNG or IFNA for fibroblasts, as detected by Northern blotting. (B) Relative real-time PCR of IRF1, ISG15, and MX1, and cDNAs from EBV-transformed B cells derived from a healthy control (C), three patients under study (P1, P2, P3), and a patient with recessive complete STAT1 deficiency (P5) homozygous for the 1928insA STAT1 mutation or from parental fibrosarcoma cell line (2C4) and STAT1-deficient U3C fibrosarcoma cell clones, untransfected (U3C) or stably cotransfected with a zeocin-resistance vector and a vector containing a mock, WT, E320Q, Q463H, or L706S STAT1 allele stimulated or not stimulated with 105 IU/ml of IFNG or IFNA for 1 h and 2 h for EBV-transformed B cells and fibroblasts, respectively, for IRF1, and for 6 h for both cellular types for ISG15 and MX1. Means values of duplicates of one experiment are shown with their respective standard variations.
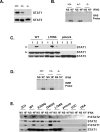
Figure 8. Mechanism of Dominance of the STAT1 Alleles for GAS-Binding Activity and of Recessivity of the STAT1 Alleles for ISRE-Binding Activity
(A) Western blot of total protein extracts (100 μg) from unstimulated EBV-transformed B-cell lines from a healthy control (+/+), P4′s father (+/−) (heterozygous for the loss-of-expression, loss-of-function STAT1 1758_1759delAG allele), and P5 (−/−) (homozygous for the loss-of-expression, loss-of-function STAT1 1928insA allele), using antibodies specific for STAT1 and STAT3. (B) EMSA of nuclear extracts (5 μg) from EBV-transformed B cells derived from a healthy control (+/+), P5 (−/−) (homozygous for the loss-of-expression, loss-of-function STAT1 1928insA allele), and P4′s father (+/−) (heterozygous for the loss-of-expression, loss-of-function STAT1 1758_1759delAG allele). EBV-transformed B cells were not stimulated (NS) or were stimulated for 30 min with 105 IU/ml IFNG. A radiolabeled GAS (FCGR1) probe was used. (C) (a) Whole-cell extracts of 107 STAT1-deficient U3C fibrosarcoma cell clones stably cotransfected with a zeocin-resistance vector and a vector containing a mock (pmock), WT, or L706S STAT1 allele were subjected to immunoprecipitation with the following biotinylated peptides: TSFGYDKPHVLV (1), corresponding to the intracellular part of IFNGR1 around the unphosphorylated Tyr-440 residue (Y); TSFG(pTyr)DKPHVLV (2), corresponding to the intracellular part of IFNGR1 around the phosphorylated Tyr-440 residue (pTyr); and SLIG(pTyr)RPTEDSK (3), corresponding to an irrelevant peptide similar to peptide 2. (b) 20 μL of each extract was taken before immunoprecipitation, and Western blotting was performed with STAT1- and STAT3-specific antibodies. (D) EMSA of nuclear extracts (5 μg) from EBV-transformed B cells derived from a healthy control (C), P4′s father (+/−) (heterozygous for the loss-of-expression, loss-of-function STAT1 1758_1759delAG allele) and P5 (−/−) (homozygous for the loss-of-expression, loss-of-function STAT1 1928insA allele). EBV-transformed B cells were not stimulated (NS) or were stimulated for 30 min with 105 IU/ml of IFNA. A radiolabeled ISRE probe was used. (E) Immunoprecipitation with a STAT1-specific antibody, followed by Western blotting with Tyr701-phospho-STAT1–specific, Tyr690-phospho-STAT2–specific, STAT1-specific, and STAT2-specific antibodies, of total protein extracts (1 mg) from a parental fibrosarcoma cell line (2C4), a STAT2-deficient U6A fibrosarcoma cell line, and STAT1-deficient U3C fibrosarcoma cell clones, untransfected (U3C) or stably cotransfected with a zeocin-resistance vector and a vector containing a mock, WT, E320Q, Q463H, or L706S STAT1 allele. The cells were not stimulated (NS) or were stimulated for 30 min with 105 IU/ml IFNA. For (B–E), one experiment representative of two independent experiments is shown.
Figure 9. Impact of Mutant STAT1 Alleles on IFNG- and IFNA-Mediated Immunity
(A) Cytokine production in the supernatant of whole blood from healthy controls (C) and patients (P1′s mother, P2, and P3) and their respective “travel” control, not stimulated (NS) or stimulated for 72 h with live BCG alone or BCG plus IL12 or IFNG. The levels of IFNG and IL12 in the supernatant were determined by enzyme-linked immunosorbent assay. One experiment representative of two independent experiments is shown. (B) Skin-derived SV40-transformed fibroblasts from a healthy control (C), the three patients under study (P1, P2, P3), a parental fibrosarcoma cell line (2C4), STAT1-deficient U3C fibrosarcoma cell line (U3C), and U3C clones stably transfected with a mock (pmock), WT, E320Q, Q463H, or L706S STAT1 alleles, were infected with HSV-1 or VSV, with or without priorstimulation with IFNA (105 IU/ml) for 24 h. Viral titers were determined after 48 h of infection. Five independent experiments are shown for the patient's cells and three independent experiments are shown for sarcoma fibroblasts. Each assay is symbolized by a different character.
Similar articles
- Dominant-negative STAT1 SH2 domain mutations in unrelated patients with Mendelian susceptibility to mycobacterial disease.
Tsumura M, Okada S, Sakai H, Yasunaga S, Ohtsubo M, Murata T, Obata H, Yasumi T, Kong XF, Abhyankar A, Heike T, Nakahata T, Nishikomori R, Al-Muhsen S, Boisson-Dupuis S, Casanova JL, Alzahrani M, Shehri MA, Elghazali G, Takihara Y, Kobayashi M. Tsumura M, et al. Hum Mutat. 2012 Sep;33(9):1377-87. doi: 10.1002/humu.22113. Epub 2012 Jun 7. Hum Mutat. 2012. PMID: 22573496 Free PMC article. - Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation.
Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, Holland SM, Schreiber RD, Casanova JL. Dupuis S, et al. Science. 2001 Jul 13;293(5528):300-3. doi: 10.1126/science.1061154. Science. 2001. PMID: 11452125 - Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency.
Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, Al Ghonaium A, Tufenkeji H, Frayha H, Al-Gazlan S, Al-Rayes H, Schreiber RD, Gresser I, Casanova JL. Dupuis S, et al. Nat Genet. 2003 Mar;33(3):388-91. doi: 10.1038/ng1097. Epub 2003 Feb 18. Nat Genet. 2003. PMID: 12590259 - Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion.
Dupuis S, Döffinger R, Picard C, Fieschi C, Altare F, Jouanguy E, Abel L, Casanova JL. Dupuis S, et al. Immunol Rev. 2000 Dec;178:129-37. doi: 10.1034/j.1600-065x.2000.17810.x. Immunol Rev. 2000. PMID: 11213797 Review.
Cited by
- Conditional Stat1 ablation reveals the importance of interferon signaling for immunity to Listeria monocytogenes infection.
Kernbauer E, Maier V, Stoiber D, Strobl B, Schneckenleithner C, Sexl V, Reichart U, Reizis B, Kalinke U, Jamieson A, Müller M, Decker T. Kernbauer E, et al. PLoS Pathog. 2012;8(6):e1002763. doi: 10.1371/journal.ppat.1002763. Epub 2012 Jun 14. PLoS Pathog. 2012. PMID: 22719255 Free PMC article. - Inherited and acquired immunodeficiencies underlying tuberculosis in childhood.
Boisson-Dupuis S, Bustamante J, El-Baghdadi J, Camcioglu Y, Parvaneh N, El Azbaoui S, Agader A, Hassani A, El Hafidi N, Mrani NA, Jouhadi Z, Ailal F, Najib J, Reisli I, Zamani A, Yosunkaya S, Gulle-Girit S, Yildiran A, Cipe FE, Torun SH, Metin A, Atikan BY, Hatipoglu N, Aydogmus C, Kilic SS, Dogu F, Karaca N, Aksu G, Kutukculer N, Keser-Emiroglu M, Somer A, Tanir G, Aytekin C, Adimi P, Mahdaviani SA, Mamishi S, Bousfiha A, Sanal O, Mansouri D, Casanova JL, Abel L. Boisson-Dupuis S, et al. Immunol Rev. 2015 Mar;264(1):103-20. doi: 10.1111/imr.12272. Immunol Rev. 2015. PMID: 25703555 Free PMC article. Review. - Genetic and Functional Identifying of Novel STAT1 Loss-of-Function Mutations in Patients with Diverse Clinical Phenotypes.
Chen X, Chen J, Chen R, Mou H, Sun G, Yang L, Jia Y, Zhao Q, Wen W, Zhou L, Ding Y, Tang X, Yang J, An Y, Zhao X. Chen X, et al. J Clin Immunol. 2022 Nov;42(8):1778-1794. doi: 10.1007/s10875-022-01339-w. Epub 2022 Aug 17. J Clin Immunol. 2022. PMID: 35976469 - Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden.
Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L. Klover PJ, et al. Neoplasia. 2010 Nov;12(11):899-905. doi: 10.1593/neo.10716. Neoplasia. 2010. PMID: 21076615 Free PMC article. - Novel STAT1 alleles in a patient with impaired resistance to mycobacteria.
Kristensen IA, Veirum JE, Møller BK, Christiansen M. Kristensen IA, et al. J Clin Immunol. 2011 Apr;31(2):265-71. doi: 10.1007/s10875-010-9480-8. Epub 2010 Nov 6. J Clin Immunol. 2011. PMID: 21057861
References
- Dorman SE, Holland SM. Interferon-gamma and interleukin-12 pathway defects and human disease. Cytokine Growth Factor Rev. 2000;11:321–333. - PubMed
- Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: The human model. Annu Rev Immunol. 2002;20:581–620. - PubMed
- Fieschi C, Bosticardo M, de Beaucoudrey L, Boisson-Dupuis S, Feinberg J, et al. A novel form of complete IL12/IL-23 receptor beta1 deficiency with cell surface-expressed nonfunctional receptors. Blood. 2004;104:2095–2101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous