Bacterial elicitation and evasion of plant innate immunity - PubMed (original) (raw)
Review
Bacterial elicitation and evasion of plant innate immunity
Robert B Abramovitch et al. Nat Rev Mol Cell Biol. 2006 Aug.
Abstract
Recent research on plant responses to bacterial attack has identified extracellular and intracellular host receptors that recognize conserved pathogen-associated molecular patterns and more specialized virulence proteins, respectively. These findings have shed light on our understanding of the molecular mechanisms by which bacteria elicit host defences and how pathogens have evolved to evade or suppress these defences.
Conflict of interest statement
Competing interests statement
The authors declare no competing financial interests.
Figures
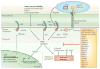
Figure 1. Disease symptoms caused by some bacterial pathogens of plants and representative virulence mechanisms used by these pathogens
Top panels (left to right): bacterial speck of tomato caused by Pseudomonas syringae pathovar (pv.) tomato; crown gall of grape caused by Agrobacterium tumefaciens; blackleg of potato caused by Erwinia carotovora subspecies atroseptica; and bacterial wilt of tomato caused by Ralstonia solanacearum. Bottom panels (left to right): P. syringae pv. tomato enters the leaf apoplastic space through stomata or wounds, and uses a type III secretion system to inject a large number of virulence (effector) proteins into the plant cell. Agrobacterium tumefaciens uses a type IV secretion system to inject a tumour-inducing transfer DNA (tDNA) into the plant cell cytoplasm. This tDNA is integrated into the plant genome and leads to the development of crown gall disease. Erwinia carotovora subspecies atroseptica uses a type II secretion system to deliver cell wall-degrading enzymes (for example, cellulases and pectinases) to the plant cell wall. Ralstonia solanacearum enters plant roots through wounds and multiplies in the xylem vessels in which it produces exopolysaccharides that are believed both to interfere with recognition and to inhibit water transport through the vascular system. Each of these four pathogens also uses other virulence mechanisms (Supplementary information 1 (table)). Ti, tumour inducing. Photo credits for top panels, left to right: G.B.M., T. Burr, A. Charkowski and P. Frey.
Figure 2. Model depicting the activation of PRR-mediated basal defences and their suppression by type III effectors
Plants possess plasma-membrane-localized pattern recognition receptors (PRRs) that consist of an extracellular leucine-rich repeat (LRR) domain and a cytoplasmic serine/threonine kinase domain. In Arabidopsis, FLS2 and elongation factor Tu (EF-Tu) receptor (EFR) are PRRs that recognize the flg22 peptide of flagellin or the elf18 peptide of EF-Tu, respectively ,,,,. Other bacterial elicitors such as lipopolysaccharide (LPS), harpins, and cold-shock protein have been identified,,,. PRR activation triggers signalling events that lead to the upregulation of over 300 plant genes,,,. A complete mitogen-activated protein kinase (MAPK) pathway and several WRKY transcription factors that function downstream of FLS2 and induce the expression of genes such as FRK1 and NHO1 have been identified. Phenotypes that are associated with activated basal defences include cell wall fortifications and the production of reactive oxygen species (ROS) and nitrogen species (NO). Delivery of effector proteins through the type III secretion system (T3SS) into plant cells is one strategy that is used by bacteria to suppress PRR-mediated defences. As many as 16 effectors have been identified that suppress basal defences,–,. The model highlights the effector AvrPto that is required for suppressing the recognition of flg22 and other pathogen-associated molecular patterns (PAMPs),. In Arabidopsis, AvrPto functions upstream of MAPK kinase kinase (MAPKKK), indicating that it targets components that function early on in the PRR pathway. The residue I96, which resides within an extended Ω-loop, is required for the basal defence suppression of AvrPto in Arabidopsis. See text for more details.
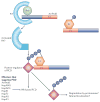
Figure 3. Suppression of R-protein-mediated defences by type III effectors
Plants have evolved resistance (R) proteins to detect the presence of type III effectors and to signal hypersensitive response (HR)-based defences in response to effectors. Several effectors have been shown to suppress HR-based defences in plants,,,,. Here we present a speculative model of how AvrPtoB suppresses HR-based defences. In tomato, the AvrPtoB N terminus is recognized by the R protein Rsb in a Prf-dependent manner to signal the HR. The AvrPtoB C terminus encodes E3 ubiquitin (Ub) ligase activity,, which includes a conserved E2 Ub-conjugating enzyme binding site. AvrPtoB might function as a scaffold to bind both to a tomato E2 Ub-conjugating enzyme and a positive regulator of HR-based programmed cell death (PCD). The E2 Ub-conjugating enzyme might then ubiquitylate the substrate to target it for degradation or alter the substrate’s localization, therefore interfering with HR-based signalling.
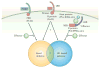
Figure 4. Factors that influence the outcome of plant–bacteria interactions
The outcome of plant–pathogen interactions might be dependent on the complement of four important factors: pathogen-associated molecular patterns (PAMPS); pattern recognition receptors (PRRs); resistance (R) proteins; and effectors. If the host can recognize the PAMPS or effectors of the pathogen, then non-host or hypersensitive response (HR)-based resistance might be elicited. If the pathogen can vary PAMPS to avoid detection or has the correct complement of effectors to suppress PRR- or R-protein-mediated resistance then disease might be observed. LRR, leucine-rich repeats; NBS, nucleotide-binding site.
Similar articles
- Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens.
Boller T, He SY. Boller T, et al. Science. 2009 May 8;324(5928):742-4. doi: 10.1126/science.1171647. Science. 2009. PMID: 19423812 Free PMC article. - Innate immunity in plants: a continuum of layered defenses.
da Cunha L, McFall AJ, Mackey D. da Cunha L, et al. Microbes Infect. 2006 Apr;8(5):1372-81. doi: 10.1016/j.micinf.2005.12.018. Epub 2006 Mar 23. Microbes Infect. 2006. PMID: 16697674 Review. - Recognition of bacterial plant pathogens: local, systemic and transgenerational immunity.
Henry E, Yadeta KA, Coaker G. Henry E, et al. New Phytol. 2013 Sep;199(4):908-15. doi: 10.1111/nph.12214. Epub 2013 Mar 20. New Phytol. 2013. PMID: 23909802 Free PMC article. Review. - Stomatal Defense a Decade Later.
Melotto M, Zhang L, Oblessuc PR, He SY. Melotto M, et al. Plant Physiol. 2017 Jun;174(2):561-571. doi: 10.1104/pp.16.01853. Epub 2017 Mar 24. Plant Physiol. 2017. PMID: 28341769 Free PMC article. Review. - Elicitation and suppression of microbe-associated molecular pattern-triggered immunity in plant-microbe interactions.
He P, Shan L, Sheen J. He P, et al. Cell Microbiol. 2007 Jun;9(6):1385-96. doi: 10.1111/j.1462-5822.2007.00944.x. Epub 2007 Apr 19. Cell Microbiol. 2007. PMID: 17451411 Review.
Cited by
- Systemic resistance induced by Bacillus lipopeptides in Beta vulgaris reduces infection by the rhizomania disease vector Polymyxa betae.
Desoignies N, Schramme F, Ongena M, Legrève A. Desoignies N, et al. Mol Plant Pathol. 2013 May;14(4):416-21. doi: 10.1111/mpp.12008. Epub 2012 Dec 19. Mol Plant Pathol. 2013. PMID: 23279057 Free PMC article. - Network properties of robust immunity in plants.
Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Tsuda K, et al. PLoS Genet. 2009 Dec;5(12):e1000772. doi: 10.1371/journal.pgen.1000772. Epub 2009 Dec 11. PLoS Genet. 2009. PMID: 20011122 Free PMC article. - Heterologous production and characterization of a pyomelanin of Antarctic Pseudomonas sp. ANT_H4: a metabolite protecting against UV and free radicals, interacting with iron from minerals and exhibiting priming properties toward plant hairy roots.
Styczynski M, Rogowska A, Nyabayo C, Decewicz P, Romaniuk F, Pączkowski C, Szakiel A, Suessmuth R, Dziewit L. Styczynski M, et al. Microb Cell Fact. 2022 Dec 16;21(1):261. doi: 10.1186/s12934-022-01990-3. Microb Cell Fact. 2022. PMID: 36527127 Free PMC article. - Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems.
Angot A, Vergunst A, Genin S, Peeters N. Angot A, et al. PLoS Pathog. 2007 Jan;3(1):e3. doi: 10.1371/journal.ppat.0030003. PLoS Pathog. 2007. PMID: 17257058 Free PMC article. Review. - Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site--leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes.
Shang J, Tao Y, Chen X, Zou Y, Lei C, Wang J, Li X, Zhao X, Zhang M, Lu Z, Xu J, Cheng Z, Wan J, Zhu L. Shang J, et al. Genetics. 2009 Aug;182(4):1303-11. doi: 10.1534/genetics.109.102871. Epub 2009 Jun 8. Genetics. 2009. PMID: 19506306 Free PMC article.
References
- Genin S, Boucher C. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu Rev Phytopathol. 2004;42:107–134. - PubMed
- Nino-Liu DO, Ronald PR, Bogdanove AJ. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. (in the press) - PubMed
- Preston GM. Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol Plant Pathol. 2000;1:263–275. - PubMed
- Van Sluys MA, et al. Comparative genomic analysis of plant-associated bacteria. Annu Rev Phytopathol. 2002;40:169–189. - PubMed
- Agrios GN. Plant Pathology. Academic Press; San Diego: 1997.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources