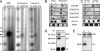MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis - PubMed (original) (raw)
MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis
Shane T Grivna et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Noncoding small RNAs have emerged as important regulators of gene expression at both transcriptional and posttranscriptional levels. Particularly, microRNA (miRNA)-mediated translational repression involving PIWI/Argonaute family proteins has been widely recognized as a novel mechanism of gene regulation. We previously reported that MIWI, a murine PIWI family member, is required for initiating spermiogenesis, a process that transforms round spermatids into mature sperm. MIWI is a cytoplasmic protein present in spermatocytes and round spermatids, and it is required for the expression of its target mRNAs involved in spermiogenesis. Most recently, we discovered a class of noncoding small RNAs called PIWI-interacting RNAs (piRNAs) that are abundantly expressed during spermiogenesis in a MIWI-dependent fashion. Here, we show that MIWI associates with both piRNAs and mRNAs in cytosolic ribonucleoprotein and polysomal fractions. As polysomes increase in early spermiogenesis, MIWI increases in polysome fractions. Moreover, MIWI associates with the mRNA cap-binding complex. Interestingly, MIWI is required for the expression of not only piRNAs but also a subset of miRNAs, despite the presence of Dicer. These results suggest that MIWI has a complicated role in the biogenesis and/or maintenance of two distinct types of small RNAs. Together, our results indicate that MIWI, a PIWI subfamily protein, uses piRNA as the major, but not exclusive, binding partner, and it is associated with translational machinery.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
Fig. 1.
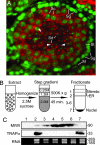
MIWI is present in both the chromatoid body and the cytosol. (A) Cross-section of a 24-days postpartum (dpp) seminiferous tubule costained for MIWI (red) and DNA (green). In a seminiferous tubule, spermatogonia (Sg) contain DAPI-intense nuclei, and they reside in the basal layer where MIWI expression is absent. Interspersed among the spermatogonia are a small number of somatic supporting cells called Sertoli cells (St), which also reside in the basal layer. The nuclei of Sertoli cells show diffuse DAPI staining but contain two distinct nucleoli (DAPI bright spots). Spermatogonia divide to produce primary spermatocytes (Sc) that contain the largest nuclei and reside mostly in the two to three subbasal layers. These primary spermatocytes are at the prophase of meiosis I. Completion of meiosis then generates round spermatids (Sd) that contain small nuclei and reside in the luminal layers. The round spermatids each contain a chromatoid body that accumulates a high concentration of MIWI (arrowheads). Round spermatids then undergo spermiogenesis to produce mature sperm (not shown). Surrounding the tubule are thin myoid cells (M), which are separated from the tubule proper by basal lamina (not shown). (Scale bar, 20 μm.) (B) Membrane-flotation assay. Testicular extract was centrifuged on a sucrose step gradient for isopycnic separation of membranes from cytosol. rER, rough endoplasmic reticulum. (C) Western blots of individual sedimentation fractions probed for MIWI and TRAPα. RNA was isolated from 20% of each fraction.
Fig. 2.
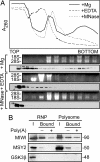
MIWI associates with mRNA in both RNPs and polysomes. (A) Untreated (+Mg) and EDTA-treated (+EDTA) or micrococcal nuclease-treated (+MNase) postnuclear testicular extract was fractionated on 15–50% sucrose density gradients and analyzed by UV spectrometry and Western blotting. RNA was isolated from 20% of every other fraction. Five percent of each fraction was immunoblotted for MIWI. (B) RNP and polysome fractions from sucrose gradients were subjected to RNP-capture assay with oligo(dT)-cellulose. Poly(A) sequences were added as a competitor. Twenty-five percent of the input and bound proteins were immunoblotted for MIWI, MSY2, and glycogen synthase kinase 3β (GSK3β).
Fig. 3.
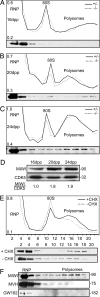
MIWI association with polysomes corresponds to increased translation during spermatogenesis. (A_–_C) Postnuclear testicular extracts from miwi+/− and _miwi_−/− mice at 16 dpp (A), 20 dpp (B), and 24 dpp (C) were fractionated on sucrose density gradients and analyzed by UV spectrometry. Numbers on the left in each panel correspond to _A_260 maximum and minimum. Five percent of every other fraction was immunoblotted for MIWI, which is shown below the corresponding _A_260 profile. (D) MIWI expression at 16, 20, and 24 dpp, normalized against CDK5 and set with the expression level at 16 dpp as 1. (E) Effect of cycloheximide (CHX) on polysome profile and MIWI distribution in cultured seminiferous tubules. (F) Immunoblots of fractions from C probed for MIWI, mouse vasa homolog (MVH), and GW182.
Fig. 4.
MIWI complexes with piRNAs and Dicer, and it is required for the expression of a subset of miRNAs. (A) Gradient fractions from untreated extract (+Mg) in Fig. 2 were combined into RNP, monosomal, and polysomal pools. Immunoprecipitation from each pool was performed with MIWI antibody (αMW) or its preimmune serum (Pre). Coprecipitated RNA was isolated, end-labeled, and separated by 6 M urea/15% PAGE. piRNAs (arrowhead) were specifically pulled down with anti-MIWI from each pool. Lane M, 10-bp DNA ladder, which migrates 10% faster than RNA molecules that contain the same number of nucleotides. (B) Northern blots of miRNAs in _miwi_−/− testes. RNA isolated from the polysomal pool of fractionated 24-dpp miwi+/− and _miwi_−/− testes and 20 μg of total RNA from 24-dpp miwi+/− and _miwi_−/− testes were probed for the indicated miRNAs, with tRNA as a loading control. (C) Northern blots of total RNA from 16- and 24-dpp miwi+/− and _miwi_−/− testes probed for the indicated miRNAs, with tRNA as a loading control. (D) Immunoprecipitation (IP) of MIWI complexes from adult testicular extract by MIWI antibody or its preimmune serum. Precipitated protein was immunoblotted (IB) for MIWI (arrow) and Dicer. Arrowhead, IgG heavy chain. (E) Western blots of testicular extracts from 24-dpp miwi+/− and _miwi_−/− mice probed for MIWI and Dicer.
Fig. 5.
MIWI associates with the cap-binding complex. Adult testicular extract was subjected to 7-methyl-GTP-Sepharose and protein A–Sepharose chromatography. Free cap analog was added as a competitor; 2.5% of input and 12% of bound proteins were immunoblotted for MIWI, the cap-binding protein eIF4E, and GSK3β.
Similar articles
- A novel class of small RNAs in mouse spermatogenic cells.
Grivna ST, Beyret E, Wang Z, Lin H. Grivna ST, et al. Genes Dev. 2006 Jul 1;20(13):1709-14. doi: 10.1101/gad.1434406. Epub 2006 Jun 9. Genes Dev. 2006. PMID: 16766680 Free PMC article. - piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis.
Zhao S, Gou LT, Zhang M, Zu LD, Hua MM, Hua Y, Shi HJ, Li Y, Li J, Li D, Wang ED, Liu MF. Zhao S, et al. Dev Cell. 2013 Jan 14;24(1):13-25. doi: 10.1016/j.devcel.2012.12.006. Dev Cell. 2013. PMID: 23328397 - miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis.
Deng W, Lin H. Deng W, et al. Dev Cell. 2002 Jun;2(6):819-30. doi: 10.1016/s1534-5807(02)00165-x. Dev Cell. 2002. PMID: 12062093 - Small RNA molecules in the regulation of spermatogenesis.
He Z, Kokkinaki M, Pant D, Gallicano GI, Dym M. He Z, et al. Reproduction. 2009 Jun;137(6):901-11. doi: 10.1530/REP-08-0494. Epub 2009 Mar 24. Reproduction. 2009. PMID: 19318589 Review. - The regulatory functions of piRNA/PIWI in spermatogenesis.
Yuan ZH, Zhao YM. Yuan ZH, et al. Yi Chuan. 2017 Aug 20;39(8):683-691. doi: 10.16288/j.yczz.17-245. Yi Chuan. 2017. PMID: 28903896 Review.
Cited by
- Biogenesis and mechanism of action of small non-coding RNAs: insights from the point of view of structural biology.
Costa MC, Leitão AL, Enguita FJ. Costa MC, et al. Int J Mol Sci. 2012;13(8):10268-10295. doi: 10.3390/ijms130810268. Epub 2012 Aug 17. Int J Mol Sci. 2012. PMID: 22949860 Free PMC article. Review. - A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis.
Morgan M, Kabayama Y, Much C, Ivanova I, Di Giacomo M, Auchynnikava T, Monahan JM, Vitsios DM, Vasiliauskaitė L, Comazzetto S, Rappsilber J, Allshire RC, Porse BT, Enright AJ, O'Carroll D. Morgan M, et al. Cell Res. 2019 Mar;29(3):221-232. doi: 10.1038/s41422-018-0128-1. Epub 2019 Jan 7. Cell Res. 2019. PMID: 30617251 Free PMC article. - A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice.
Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. Nonomura K, et al. Plant Cell. 2007 Aug;19(8):2583-94. doi: 10.1105/tpc.107.053199. Epub 2007 Aug 3. Plant Cell. 2007. PMID: 17675402 Free PMC article. - Towards a Multi-Omics of Male Infertility.
Wagner AO, Turk A, Kunej T. Wagner AO, et al. World J Mens Health. 2023 Apr;41(2):272-288. doi: 10.5534/wjmh.220186. Epub 2023 Jan 4. World J Mens Health. 2023. PMID: 36649926 Free PMC article. Review. - Unraveling mitochondrial piRNAs in mouse embryonic gonadal cells.
Barreñada O, Larriba E, Fernández-Pérez D, Brieño-Enríquez MÁ, Del Mazo Martínez J. Barreñada O, et al. Sci Rep. 2022 Jun 24;12(1):10730. doi: 10.1038/s41598-022-14414-4. Sci Rep. 2022. PMID: 35750721 Free PMC article.
References
- Kozak M. Annu. Rev. Cell Biol. 1992;8:197–225. - PubMed
- Storz G., Altuvia S., Wassarman K. M. Annu. Rev. Biochem. 2005;74:199–217. - PubMed
- Wilusz C. J., Wilusz J. Trends Genet. 2004;20:491–497. - PubMed
- Kleene K. C. Cytogenet. Genome Res. 2003;103:217–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA59365/CA/NCI NIH HHS/United States
- HD42012/HD/NICHD NIH HHS/United States
- T32 CA059365/CA/NCI NIH HHS/United States
- R01 HD042012/HD/NICHD NIH HHS/United States
- R37 HD042012/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous