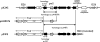Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate - PubMed (original) (raw)
Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate
Ying-Tsong Chen et al. Antimicrob Agents Chemother. 2006 Nov.
Abstract
A plasmid containing the qnrS quinolone resistance determinant and the gene encoding the SHV-2 beta-lactamase has been discovered from a clinical Klebsiella pneumoniae strain isolated in Taiwan. The complete 98-kb sequence of this plasmid, designated pK245, was determined by using a whole-genome shotgun approach. Transfer of pK245 conferred low-level resistance to fluoroquinolones in electroporant Escherichia coli epi300. The sequence of the immediate region surrounding qnrS in pK245 is nearly identical (>99% identity) to those of pAH0376 from Shigella flexneri and pINF5 from Salmonella enterica serovar Infantis, the two other qnrS-carrying plasmids reported to date, indicating a potential common origin. Other genes conferring resistance to aminoglycosides (aacC2, strA, and strB), chloramphenicol (catA2), sulfonamides (sul2), tetracycline (tetD), and trimethoprim (dfrA14) were also detected in pK245. The dfrA14 gene is carried on a class I integron. Several features of this plasmid, including three separate regions containing putative replicons, a partitioning-control system, and a type II restriction modification system, suggest that it may be able to replicate and adapt in a variety of hosts. Although no critical conjugative genes were detected, multiple insertion sequence elements were found scattered throughout pK245, and these may facilitate the dissemination of the antimicrobial resistance determinants. We conclude that pK245 is a chimera which acquired its multiple antimicrobial resistance determinants horizontally from different sources. The identification of pK245 plasmid expands the repertoire of the coexistence of quinolone and extended-spectrum-beta-lactam resistance determinants in plasmids carried by various species of the family Enterobacteriaceae in different countries.
Figures
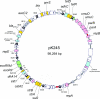
FIG. 1.
Circular map of plasmid pK245. Genes are color coded as follows: yellow, antimicrobial resistance associated; pink, plasmid replication and partitioning; black (with white triangles indicating their orientation), IS_26_; different shades of gray, other IS elements; green, nuclease and methylase; light blue, other functions; white, unknown functions. The arrows on the ORFs indicate the direction of transcription. Iteron regions are marked red.
FIG. 2.
Schematic diagram comparing the qnrS_-containing region of pK245 with those of pAH0376 from S. flexneri and pINF5 from S. enterica serovar Infantis. The ORFs are shown as arrows, with the arrowhead indicating the direction of transcription. A 12.4-kb region of pK245, which is flanked by two copies of IS_26, is illustrated on the top. The terminal inverted repeats on both sides of IS_26_ are shown in triangles. The Tn_3_ element is absent from pK245, and the deduced amino acid sequence of bla from pK245 is 61% identical and 75% similar to the TEM-1 β-lactamase. The 2.6-kb and 5.7-kb regions in pK245 that are similar to pAH0376 and pINF5, respectively, are also indicated by vertical lines. The stippled arrows indicate the qnrS gene. Black arrows are hypothetical ORFs. Although some predictions of the putative ORFs in these three plasmids are different, the nucleotide sequences of the homologous regions are more than 99.8% identical.
Similar articles
- qnrA prevalence in extended-spectrum beta-lactamase-positive Enterobacteriaceae isolates from Turkey.
Oktem IM, Gulay Z, Bicmen M, Gur D; HITIT Project Study Group. Oktem IM, et al. Jpn J Infect Dis. 2008 Jan;61(1):13-7. Jpn J Infect Dis. 2008. PMID: 18219128 - Association of QnrB determinants and production of extended-spectrum beta-lactamases or plasmid-mediated AmpC beta-lactamases in clinical isolates of Klebsiella pneumoniae.
Pai H, Seo MR, Choi TY. Pai H, et al. Antimicrob Agents Chemother. 2007 Jan;51(1):366-8. doi: 10.1128/AAC.00841-06. Epub 2006 Oct 30. Antimicrob Agents Chemother. 2007. PMID: 17074790 Free PMC article. - Identification of the plasmid-borne quinolone resistance gene qnrS in Salmonella enterica serovar Infantis.
Kehrenberg C, Friederichs S, de Jong A, Michael GB, Schwarz S. Kehrenberg C, et al. J Antimicrob Chemother. 2006 Jul;58(1):18-22. doi: 10.1093/jac/dkl213. Epub 2006 May 23. J Antimicrob Chemother. 2006. PMID: 16720566 - Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae.
Nordmann P, Poirel L. Nordmann P, et al. J Antimicrob Chemother. 2005 Sep;56(3):463-9. doi: 10.1093/jac/dki245. Epub 2005 Jul 14. J Antimicrob Chemother. 2005. PMID: 16020539 Review. - Plasmid-mediated quinolone resistance--current knowledge and future perspectives.
Guan X, Xue X, Liu Y, Wang J, Wang Y, Wang J, Wang K, Jiang H, Zhang L, Yang B, Wang N, Pan L. Guan X, et al. J Int Med Res. 2013 Feb;41(1):20-30. doi: 10.1177/0300060513475965. Epub 2013 Jan 23. J Int Med Res. 2013. PMID: 23569126 Review.
Cited by
- Epigenetic Influence of Dam Methylation on Gene Expression and Attachment in Uropathogenic Escherichia coli.
Stephenson SA, Brown PD. Stephenson SA, et al. Front Public Health. 2016 Jun 27;4:131. doi: 10.3389/fpubh.2016.00131. eCollection 2016. Front Public Health. 2016. PMID: 27446897 Free PMC article. - Characterization of Four Multidrug Resistance Plasmids Captured from the Sediments of an Urban Coastal Wetland.
Botts RT, Apffel BA, Walters CJ, Davidson KE, Echols RS, Geiger MR, Guzman VL, Haase VS, Montana MA, La Chat CA, Mielke JA, Mullen KL, Virtue CC, Brown CJ, Top EM, Cummings DE. Botts RT, et al. Front Microbiol. 2017 Oct 10;8:1922. doi: 10.3389/fmicb.2017.01922. eCollection 2017. Front Microbiol. 2017. PMID: 29067005 Free PMC article. - Transferable Plasmids of Salmonella enterica Associated With Antibiotic Resistance Genes.
McMillan EA, Jackson CR, Frye JG. McMillan EA, et al. Front Microbiol. 2020 Oct 8;11:562181. doi: 10.3389/fmicb.2020.562181. eCollection 2020. Front Microbiol. 2020. PMID: 33133037 Free PMC article. Review. - First report of Klebsiella quasipneumoniae harboring _bla_KPC-2 in Saudi Arabia.
Hala S, Antony CP, Alshehri M, Althaqafi AO, Alsaedi A, Mufti A, Kaaki M, Alhaj-Hussein BT, Zowawi HM, Al-Amri A, Pain A. Hala S, et al. Antimicrob Resist Infect Control. 2019 Dec 19;8:203. doi: 10.1186/s13756-019-0653-9. eCollection 2019. Antimicrob Resist Infect Control. 2019. PMID: 31890159 Free PMC article. - qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin.
Cavaco LM, Hasman H, Xia S, Aarestrup FM. Cavaco LM, et al. Antimicrob Agents Chemother. 2009 Feb;53(2):603-8. doi: 10.1128/AAC.00997-08. Epub 2008 Nov 24. Antimicrob Agents Chemother. 2009. PMID: 19029321 Free PMC article.
References
- Barthélémy, M., J. Peduzzi, H. B. Yaghlane, and R. Labia. 1988. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 231:217-220. - PubMed
- Chen, Y. T., H. Y. Chang, Y. C. Lai, C. C. Pan, S. F. Tsai, and H. L. Peng. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337:189-198. - PubMed
- Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. Clinical and Laboratory Standards Institute, Wayne, Pa.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases