Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant - PubMed (original) (raw)
Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant
Gabriele Bönemann et al. Antimicrob Agents Chemother. 2006 Sep.
Abstract
Plasmid-encoded quinolone resistance was previously reported for different bacteria isolated from patients not only in the United States and Asia but also in Europe. Here we describe the isolation, by applying a new selection strategy, of the quinolone resistance plasmid pGNB2 from an activated sludge bacterial community of a wastewater treatment plant in Germany. The hypersensitive Escherichia coli strain KAM3 carrying a mutation in the multidrug efflux system genes acrAB was transformed with total plasmid DNA preparations isolated from activated sludge bacteria and subsequently selected on medium containing the fluoroquinolone norfloxacin. This approach resulted in the isolation of plasmid pGNB2 conferring decreased susceptibility to nalidixic acid and to different fluoroquinolones. Analysis of the pGNB2 nucleotide sequence revealed that it is 8,469 bp in size and has a G+C content of 58.2%. The plasmid backbone is composed of a replication initiation module (repA-repC) belonging to the IncQ-family and a two-component mobilization module that confers horizontal mobility to the plasmid. In addition, plasmid pGNB2 carries an accessory module consisting of a transposon Tn1721 remnant and the quinolone resistance gene, qnrS2, that is 92% identical to the qnrS gene located on plasmid pAH0376 from Shigella flexneri 2b. QnrS2 belongs to the pentapeptide repeat protein family and is predicted to protect DNA-gyrase activity against quinolones. This is not only the first report on a completely sequenced plasmid mediating quinolone resistance isolated from an environmental sample but also on the first qnrS-like gene detected in Europe.
Figures
FIG. 1.
Genetic map of the quinolone resistance plasmid pGNB2. Coding regions are indicated by arrows giving the direction of transcription. The plasmid is composed of the replication initiation genes repA and repC, the putative mobilization module genes orf1 and mobC, a gene of unknown function (orf2), and the accessory module genes orfI and qnrS2 conferring quinolone resistance. The origins of vegetative (oriV) and transfer replication (oriT) are marked by black circles. The next circle closer to the center represents the G+C plot, where a G+C content of <50% is shown in gray and a G+C content of >50% is shown in black. The G+C plot was generated by using the GenDB (version 2.2) tool. The inner circle gives the scale of the plasmid in base pairs. The HindIII site used for insertion of the Tn_5_ kanamycin/neomycin resistance gene cassette aphII is shown.
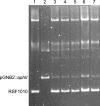
FIG. 2.
Compatibility of pGNB2 and the prototype IncQ plasmid RSF1010. E. coli JM108(RSF1010) (lane 1), E. coli DH5α(pGNB2-aphII) (lane 2), and JM108 transformants carrying both plasmids (lanes 3 to 7) were analyzed in Eckhardt gels. Plasmids RSF1010 and pGNB2::aphII are, respectively, 8,684 and 11,893 bp in size. Coexistence of RSF1010 and pGNB2::aphII in one cell could be shown for 24 randomly selected JM108 transformants.
FIG. 3.
Sequence comparison of the plasmid-encoded QnrS2 protein of pGNB2 with other Qnr proteins. QnrA1 proteins (AAL60061, AAY46799, AAY46800, AAX18268, AAX18278, AAP20926, AAP20910, and AAW31096) are encoded on plasmids from Klebsiella pneumoniae or Escherichia coli. QnrB2 is encoded on the Citrobacter koseri plasmid pMG301 (DQ351242). Although the host of the plasmid that encodes for QnrS2 is unknown, QnrS1 is encoded on a plasmid from Shigella flexneri 2b (AB187515). Dots indicate identical amino acids compared to QnrA1. Pentapeptide repeats with the consensus sequence (A/C)-(D/N)-(L/F)-X-X that occur at the same position in all Qnr-proteins are marked with asterisks. The glycine residue (G) separating the two domains of Qnr proteins is marked by a vertical arrow.
Similar articles
- Complete nucleotide sequence of a quinolone resistance gene (qnrS2) carrying plasmid of Aeromonas hydrophila isolated from fish.
Majumdar T, Das B, Bhadra RK, Dam B, Mazumder S. Majumdar T, et al. Plasmid. 2011 Jul;66(2):79-84. doi: 10.1016/j.plasmid.2011.05.001. Epub 2011 Jun 12. Plasmid. 2011. PMID: 21664930 - A small IncQ-type plasmid carrying the quinolone resistance (qnrS2) gene from Aeromonas hydrophila.
Han JE, Kim JH, Choresca CH Jr, Shin SP, Jun JW, Chai JY, Park SC. Han JE, et al. Lett Appl Microbiol. 2012 Apr;54(4):374-6. doi: 10.1111/j.1472-765X.2012.03208.x. Epub 2012 Feb 27. Lett Appl Microbiol. 2012. PMID: 22260532 - Plasmid-mediated quinolone resistance.
Martínez-Martínez L, Eliecer Cano M, Manuel Rodríguez-Martínez J, Calvo J, Pascual A. Martínez-Martínez L, et al. Expert Rev Anti Infect Ther. 2008 Oct;6(5):685-711. doi: 10.1586/14787210.6.5.685. Expert Rev Anti Infect Ther. 2008. PMID: 18847406 Review. - Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae.
Nordmann P, Poirel L. Nordmann P, et al. J Antimicrob Chemother. 2005 Sep;56(3):463-9. doi: 10.1093/jac/dki245. Epub 2005 Jul 14. J Antimicrob Chemother. 2005. PMID: 16020539 Review.
Cited by
- Molecular Characterization and Comparative Genomics of IncQ-3 Plasmids Conferring Resistance to Various Antibiotics Isolated from a Wastewater Treatment Plant in Warsaw (Poland).
Piotrowska M, Dziewit L, Ostrowski R, Chmielowska C, Popowska M. Piotrowska M, et al. Antibiotics (Basel). 2020 Sep 17;9(9):613. doi: 10.3390/antibiotics9090613. Antibiotics (Basel). 2020. PMID: 32957637 Free PMC article. - First Report of an OXA-48- and CTX-M-213-Producing Kluyvera Species Clone Recovered from Patients Admitted in a University Hospital in Madrid, Spain.
Hernández-García M, León-Sampedro R, Pérez-Viso B, Morosini MI, López-Fresneña N, Díaz-Agero C, Coque TM, Ruiz-Garbajosa P, Cantón R. Hernández-García M, et al. Antimicrob Agents Chemother. 2018 Oct 24;62(11):e01238-18. doi: 10.1128/AAC.01238-18. Print 2018 Nov. Antimicrob Agents Chemother. 2018. PMID: 30181367 Free PMC article. - Replication and conjugative mobilization of broad host-range IncQ plasmids.
Meyer R. Meyer R. Plasmid. 2009 Sep;62(2):57-70. doi: 10.1016/j.plasmid.2009.05.001. Epub 2009 May 22. Plasmid. 2009. PMID: 19465049 Free PMC article. Review. - Emergence of the quinolone resistance-mediating gene aac(6')-Ib-cr in extended-spectrum-beta-lactamase-producing Klebsiella isolates collected in Slovenia between 2000 and 2005.
Ambrozic Avgustin J, Keber R, Zerjavic K, Orazem T, Grabnar M. Ambrozic Avgustin J, et al. Antimicrob Agents Chemother. 2007 Nov;51(11):4171-3. doi: 10.1128/AAC.01480-06. Epub 2007 Sep 10. Antimicrob Agents Chemother. 2007. PMID: 17846136 Free PMC article. - Plasmid-mediated QnrS2 determinant from a clinical Aeromonas veronii isolate.
Sánchez-Céspedes J, Blasco MD, Marti S, Alba V, Alcalde E, Esteve C, Vila J. Sánchez-Céspedes J, et al. Antimicrob Agents Chemother. 2008 Aug;52(8):2990-1. doi: 10.1128/AAC.00287-08. Epub 2008 May 27. Antimicrob Agents Chemother. 2008. PMID: 18505860 Free PMC article. No abstract available.
References
- Acar, J. F., and F. W. Goldstein. 1997. Trends in bacterial resistance to fluoroquinolones. Clin. Infect. Dis. 24(Suppl. 1):S67-S73. - PubMed
- Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. - PubMed
- Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high-efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-379.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources