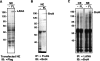Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes - PubMed (original) (raw)
Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes
Jianxin You et al. J Virol. 2006 Sep.
Abstract
The latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus (KSHV) is required for viral episome maintenance in host cells during latent infection. Two regions of the protein have been implicated in tethering LANA/viral episomes to the host mitotic chromosomes, and LANA chromosome-binding sites are subjects of high interest. Because previous studies had identified bromodomain protein Brd4 as the mitotic chromosome anchor for the bovine papillomavirus E2 protein, which tethers the viral episomes to host mitotic chromosomes (J. You, J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley, Cell 117:349-360, 2004, and J. You, M. R. Schweiger, and P. M. Howley, J. Virol. 79:14956-14961, 2005), we examined whether KSHV LANA interacts with Brd4. We found that LANA binds Brd4 in vivo and in vitro and that the binding is mediated by a direct protein-protein interaction between the ET (extraterminal) domain of Brd4 and a carboxyl-terminal region of LANA previously implicated in chromosome binding. Brd4 associates with mitotic chromosomes throughout mitosis and demonstrates a strong colocalization with LANA and the KSHV episomes on host mitotic chromosomes. Although another bromodomain protein, RING3/Brd2, binds to LANA in a similar fashion in vitro, it is largely excluded from the mitotic chromosomes in KSHV-uninfected cells and is partially recruited to the chromosomes in KSHV-infected cells. These data identify Brd4 as an interacting protein for the carboxyl terminus of LANA on mitotic chromosomes and suggest distinct functional roles for the two bromodomain proteins RING3/Brd2 and Brd4 in LANA binding. Additionally, because Brd4 has recently been shown to have a role in transcription, we examined whether Brd4 can regulate the CDK2 promoter, which can be transactivated by LANA.
Figures
FIG. 1.
KSHV LANA protein binding to Brd4. (A) Expression of FLAG-tagged LANA in C33A cells. Nuclear extracts (NE) from C33A transfected with a FLAG-LANA construct (FL) or empty vector (−) were analyzed on an SDS-PAGE gel and immunoblotted with anti-FLAG M2 antibody. Molecular masses of the markers are indicated on the left. (B) Anti-Brd4 immunoblot analysis of anti-FLAG immunoprecipitation. NE of C33A cells transfected with FLAG-LANA plasmid (FL) or empty vector (−) were immunoprecipitated with anti-FLAG M2 beads. Western blot with an anti-Brd4 rabbit polyclonal antibody, C-MCAP, demonstrated that human Brd4 protein is present in the sample coimmunoprecipitated with LANA. (C) Anti-Brd4 immunoblot analysis of cellular proteins coimmunoprecipitated with LANA. C33A cells were transfected with FLAG-LANA plasmid (FL) or empty vector (−). Cytoplasmic extracts (CE) and nuclear extracts (NE) immunoprecipitated with anti-LANA antibody LN53 were immunoblotted with the Brd4 antibody C-MCAP. IB, immunoblot.
FIG. 2.
Mapping of the LANA-binding domain on Brd4 protein. (A) Diagram of human Brd4 (HBRD4) full-length protein and the fragments used to map the LANA binding domain. The full-length Brd4 and each indicated fragment was translated and labeled by [35S]Met using in vitro transcription and translation (TNT). BD, bromodomain. (B) GST-tagged LANA constructs for testing Brd4 binding. Shown are the domain structures of LANA protein and the fragments of LANA fused to GST (LANA aa 1-23, LANA-N; LANA aa 982-1162, LANA-C). (C) In vitro binding of Brd4 to LANA protein. Each Brd4 TNT product was tested for LANA binding using GST-LANA-N (N) or GST-LANA-C (C) immobilized on glutathione resin. GST-E2TA (A) and GST-E2TR (R) beads were used as positive and negative controls, respectively. GST alone (−) was used as a negative control. Aliquots (40 μl) of sample eluted from GST fusion beads were resolved by SDS-PAGE along with 25% of the input sample (lanes I), and they were detected by autoradiography. Arrows indicate the size of full-length translation products. FL, full length.
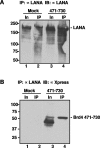
FIG. 3.
Brd4 aa 471-730 interacts with LANA in vivo. (A) Anti-LANA immunoblot detection of the input and immunoprecipitated LANA protein. Exponentially growing BJAB cells (1 × 107) expressing full-length LANA (1) were transiently transfected with either a pcDNA4C plasmid expressing His-Xpress-Brd4 aa 471-730 (471-730) or empty vector (Mock). Forty-eight hours after transfection, cells were lysed as described previously (29) and were immunoprecipitated with 1 μg of anti-LANA antibody LN53. The immunoprecipitated samples (IP) were analyzed with the input sample (In) on an SDS-8% PAGE gel and blotted with an anti-LANA polyclonal human serum to show that anti-FLAG immunoprecipitation of LANA was not affected by Brd4 aa 471-730 expression. (B) Anti-Xpress immunoblot detection of the interaction between LANA and Brd4 aa 471-730. Anti-LANA-immunoprecipitated samples were blotted with an anti-Xpress antibody. The Brd4 aa 471-730 fragment was specifically coimmunoprecipitated with LANA in the transfected cells but not in the “mock” transfection. Similar to the full-length Brd4, the Brd4 aa 471-730 fragment migrated as a protein larger than the deduced molecular mass of 26 kDa. IB, immunoblot.
FIG. 4.
Mapping of the Brd4-binding domain on the LANA protein. (A) In vitro binding of LANA fragments to Brd4 protein. LANA and each indicated LANA subfragment were translated in vitro and labeled using [35S]Met. Each TNT reaction was tested for binding using bacterially expressed GST-Brd4 protein immobilized on glutathione resin. GST alone (−) was used as a negative control. Aliquots of sample eluted from GST fusion beads were resolved by SDS-PAGE along with 20% of the input sample (lanes I) and were detected by autoradiography. (B) Summary of the interactions of various regions of LANA with GST-Brd4.
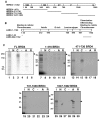
FIG. 5.
Direct interaction of Brd4 with the LANA C terminus. (A) GST-tagged Brd4 and Brd2 constructs for testing LANA binding. Shown are the domain structures of Brd4 aa 471-730 and the Brd2 ET domain region examined in this experiment. (B) In vitro direct binding of Brd4 to the LANA C terminus. Each MBP-LANA product was tested for binding on GST-Brd4 aa 471-594 (A), GST-Brd4 aa 595-730 (B), GST-Brd2 aa 592-754 (C), or GST protein alone (−) immobilized on glutathione resin. Aliquots (10 μl) of sample eluted from beads were resolved by SDS-PAGE along with 30% of the input sample (I) and were detected by Coomassie blue staining.
FIG. 6.
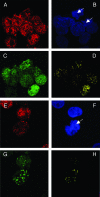
Colocalization of Brd4 and LANA on mitotic chromosomes in BJAB cells stably expressing LANA. BJAB/F-LANA cells stably expressing LANA (A to D) and BJAB/F-LANA stable cells carrying the p8TR artificial KSHV episomes (E to H) were double stained with an anti-Brd4 rabbit polyclonal antibody, N-MCAP, and the anti-LANA Rat monoclonal antibody, LN53. The staining was detected by incubation with an Alexa Fluor 594 goat anti-rabbit IgG (A and E) and a goat anti-rat IgG FITC conjugate (C and G), respectively. Cells were also counter stained with DAPI to identify nuclei and mitotic chromosomes (B and F) (the arrows indicate mitotic chromosomes). Cells were examined under a Zeiss LSM 510 UV upright confocal microscope, and the colocalized Brd4 and LANA staining was visualized using Zeiss LSM 510 software (D and H). Colocalization of Brd4 and LANA protein was distinctly observed on mitotic chromosomes as bright yellow dots (H).
FIG. 7.
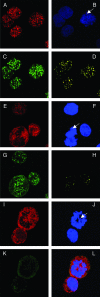
Colocalization of Brd4 and LANA on mitotic chromosomes in KSHV-positive BCLM cells. (A to D) BCLM cells were double stained with N-MCAP (A) and LN53 (C) as described in the legend to Fig. 6. Cells were also counterstained with DAPI (B) (the arrow indicates a mitotic chromosome). Colocalization of Brd4 and LANA was distinctly observed on mitotic chromosomes as bright yellow dots (D). (E to H) BCLM cells were double stained with an anti-Brd2 peptide rabbit polyclonal antibody and LN53. The staining was detected by incubation with a Alexa Fluor 594 goat anti-rabbit IgG (E) and a goat anti-rat IgG FITC conjugate (G), respectively. The staining of DAPI is shown in panel F. The overlay signal of panels E and G is shown in panel H. In contrast to the LANA staining, a majority of Brd2 staining is excluded from the mitotic chromosomes (E and H). (I to L) KSHV-negative BJAB cells were double stained for Brd2 and LANA as for panels E to H. The overlay signal is shown in panel L. In contrast to the status in KSHV-positive cells, the Brd2 staining is almost completely excluded from mitotic chromosomes.
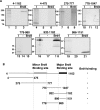
FIG. 8.
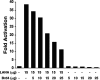
Brd4 downregulates the CDK2 promoter and LANA transactivation of the CDK2 promoter. BJAB B-lymphoma cells were transfected with the indicated plasmids. Thirty-six hours posttransfection, luciferase activity was determined and compared to the activity after transfection with reporter alone to determine the fold activation or repression. The experiment shown is representative of more than 10 experiments. The reduction in activation was not due to diminished LANA expression, as determined by Western blotting.
Similar articles
- Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest.
Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. Ottinger M, et al. J Virol. 2006 Nov;80(21):10772-86. doi: 10.1128/JVI.00804-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928766 Free PMC article. - Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1.
Viejo-Borbolla A, Ottinger M, Brüning E, Bürger A, König R, Kati E, Sheldon JA, Schulz TF. Viejo-Borbolla A, et al. J Virol. 2005 Nov;79(21):13618-29. doi: 10.1128/JVI.79.21.13618-13629.2005. J Virol. 2005. PMID: 16227282 Free PMC article. - Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes.
You J, Croyle JL, Nishimura A, Ozato K, Howley PM. You J, et al. Cell. 2004 Apr 30;117(3):349-60. doi: 10.1016/s0092-8674(04)00402-7. Cell. 2004. PMID: 15109495 - KSHV Genome Replication and Maintenance.
Purushothaman P, Dabral P, Gupta N, Sarkar R, Verma SC. Purushothaman P, et al. Front Microbiol. 2016 Feb 1;7:54. doi: 10.3389/fmicb.2016.00054. eCollection 2016. Front Microbiol. 2016. PMID: 26870016 Free PMC article. Review. - Structure and function of latency-associated nuclear antigen.
Verma SC, Lan K, Robertson E. Verma SC, et al. Curr Top Microbiol Immunol. 2007;312:101-36. doi: 10.1007/978-3-540-34344-8_4. Curr Top Microbiol Immunol. 2007. PMID: 17089795 Free PMC article. Review.
Cited by
- Replication-dependent and independent mechanisms for the chromosome-coupled persistence of a selfish genome.
Liu YT, Chang KM, Ma CH, Jayaram M. Liu YT, et al. Nucleic Acids Res. 2016 Sep 30;44(17):8302-23. doi: 10.1093/nar/gkw694. Epub 2016 Aug 4. Nucleic Acids Res. 2016. PMID: 27492289 Free PMC article. - Kaposi's sarcoma-associated herpesvirus LANA protein downregulates nuclear glycogen synthase kinase 3 activity and consequently blocks differentiation.
Liu J, Martin H, Shamay M, Woodard C, Tang QQ, Hayward SD. Liu J, et al. J Virol. 2007 May;81(9):4722-31. doi: 10.1128/JVI.02548-06. Epub 2007 Feb 21. J Virol. 2007. PMID: 17314169 Free PMC article. - Function of BCLAF1 in human disease.
Yu Z, Zhu J, Wang H, Li H, Jin X. Yu Z, et al. Oncol Lett. 2022 Feb;23(2):58. doi: 10.3892/ol.2021.13176. Epub 2021 Dec 22. Oncol Lett. 2022. PMID: 34992690 Free PMC article. Review. - Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4.
Lin YJ, Umehara T, Inoue M, Saito K, Kigawa T, Jang MK, Ozato K, Yokoyama S, Padmanabhan B, Güntert P. Lin YJ, et al. Protein Sci. 2008 Dec;17(12):2174-9. doi: 10.1110/ps.037580.108. Epub 2008 Sep 24. Protein Sci. 2008. PMID: 18815416 Free PMC article. - KSHV LANA--the master regulator of KSHV latency.
Uppal T, Banerjee S, Sun Z, Verma SC, Robertson ES. Uppal T, et al. Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
References
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
- Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856-861. - PubMed
- Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA116720/CA/NCI NIH HHS/United States
- R03 CA128006/CA/NCI NIH HHS/United States
- R01 CA 116720/CA/NCI NIH HHS/United States
- R01 CA082036/CA/NCI NIH HHS/United States
- P01 CA 050661/CA/NCI NIH HHS/United States
- P01 CA050661/CA/NCI NIH HHS/United States
- CA 082036/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous