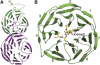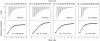Structural basis for molecular recognition and presentation of histone H3 by WDR5 - PubMed (original) (raw)
Structural basis for molecular recognition and presentation of histone H3 by WDR5
Anja Schuetz et al. EMBO J. 2006.
Abstract
Histone methylation at specific lysine residues brings about various downstream events that are mediated by different effector proteins. The WD40 domain of WDR5 represents a new class of histone methyl-lysine recognition domains that is important for recruiting H3K4 methyltransferases to K4-dimethylated histone H3 tail as well as for global and gene-specific K4 trimethylation. Here we report the crystal structures of full-length WDR5, WDR5Delta23 and its complexes with unmodified, mono-, di- and trimethylated histone H3K4 peptides. The structures reveal that WDR5 is able to bind all of these histone H3 peptides, but only H3K4me2 peptide forms extra interactions with WDR5 by use of both water-mediated hydrogen bonding and the altered hydrophilicity of the modified lysine 4. We propose a mechanism for the involvement of WDR5 in binding and presenting histone H3K4 for further methylation as a component of MLL complexes.
Figures
Figure 1
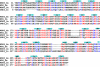
Structure-based sequence alignment of WDR5 homologs. Identical residues are colored in red and conserved residues are colored in blue. The secondary structural elements of human WDR5 are shown above the sequence. Hs: Homo sapiens; Dm: Drosophila melanogaster; Ce: Caenorhabditis elegans; Sc: Saccharomyces cerevisiae.
Figure 2
Overall structures of His-tagged WDR5Δ23 alone and in complex with H3K4me2. (A) The N-terminal tail of His-tagged WDR5Δ23 binds to the top face of the β-propeller core domain of a symmetry-related molecule. The structure of full-length WDR5 is similar to that of His-tagged WDR5Δ23. (B) The K4-dimethylated H3 peptide binds to the top face of WDR5 with Arg2 inserting into the central channel of the β-propeller structure. The seven blades of the WD40 domain are numbered from 1 to 7, and the strands of each blade are numbered A–D from the innermost strand to the outermost strand.
Figure 3
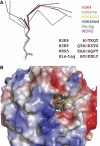
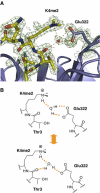
Structures of WDR5 bound to different peptides. (A) Superimposition of the six peptides that are bound to WDR5 core domain. Unmodified histone H3 is colored in red, H3K4me is colored in green, H3K4me2 is colored in yellow, H3K4me3 is colored in blue, a stretch of residues from the His-tag of WDR5Δ23 is colored in cyan and the N-terminal tail (aa11–18) of WDR5 is colored in magenta. The arginine-containing sequences of histone H3K4, H3K9, WDR5 N-terminal tail (aa11–18) and part of the His-tag from WDR5Δ23 that binds to WDR5 core domain are also shown in this panel. (B) Surface representation of WDR5 core domain. H3K4me2 peptide is colored in yellow, of which Thr3 and K4me2 are shown in a stick model. His-tag fragment of WDR5Δ23 is colored in cyan and the glutamic acid corresponding to Thr3 in H3 is shown in a stick model.
Figure 4
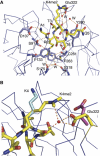
Interaction between WDR5 and histone peptides. (A) H3K4me2 peptide is shown in a stick model colored in yellow, and residues in WDR5 that make hydrogen bonds with H3K4me2 are also shown in a stick model and colored in blue. Hydrogen bonds are denoted as orange dotted lines. (B) Interaction of different states of H3K4 with Glu322 in WDR5. Dimethylated K4 is shown as a stick model colored in yellow, which interacts with Glu322 in WDR5, colored in yellow, via a water molecule. Unmodified K4 in the WDR5–H3K4 complex is colored in cyan, which is stabilized by crystal contacts. The side chain of Glu322 in this complex is either disordered or points to solvent. In the WDR5Δ23–H3K4me complex structure, the side chain of K4me is disordered. The side chain of Glu322 is either disordered or adopts the conformation of Glu322 in the WDR5Δ23–H3K4me2 complex. K4me3 in the WDR5–H3K4me3 complex is disordered, and Glu322, colored in magenta, points to solvent.
Figure 5
Detailed interaction between me2K4 in H3K4me2 and Glu322 in WDR5. (A) The H3K4me2 peptide and WDR5 are colored in yellow and blue, respectively. The electron density map is contoured at 1σ. Potential water-mediated hydrogen bonds are shown as dashed orange lines. (B) The lower panel shows schematics of two alternative hydrogen binding networks involving the water molecule.
Figure 6
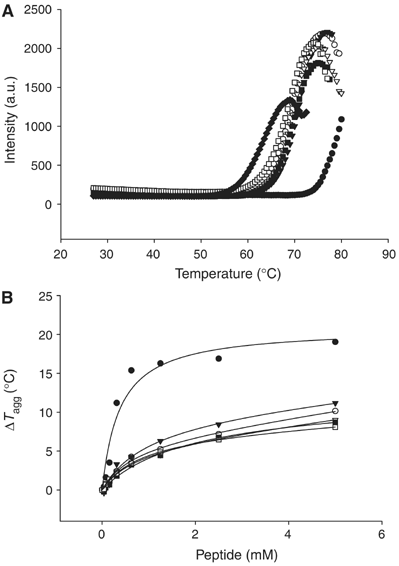
Effects of histone peptides on the thermostability of WDR5Δ23. (A) Thermostability of WDR5Δ23 alone (♦), and in the presence of 2.5 mM of H3K4me1 (aa1–11) (□), H3K4me2 (aa1–11) (•), H3K4me3 (aa1–11) (○), H3K4 (aa1–11) (▵), WDR5 (aa11–18) (▾) and H3K9me3 (▪). (B) Thermostability of WDR5Δ23 as a function of peptide concentration.
Figure 7
Isothermal titration calorimetry data for binding of methylated and unmodified H3 peptides to WDR5 at 25°C. The upper panels show the sequential heat pulses for peptide–WDR5 binding and the lower panels show the integrated heat data, corrected for the heat of dilution and fit to a single-site binding model using Origin Software. The error values are the s.d. of the values of two titration experiments.
Figure 8
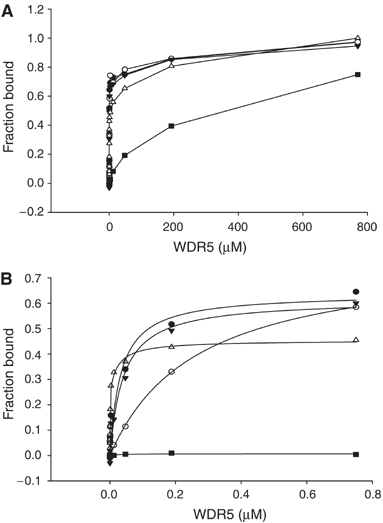
Fluorescence polarization measurements of peptide binding to WDR5. (A) Different concentrations of WDR5 were added to 30 nM of fluorescein-labeled peptide, H3K4me3 (aa1–11) (•), H3K4me2 (aa1–11) (○), H3K4me1 (aa1–8) (▾), unmodified H3K4 (aa1–11) (▵) and R2A H3 (aa1–8) (▪). Points are connected by straight lines. (B) The data measured up to 0.8 μM WDR5 fit to a one-site binding model.
Similar articles
- Molecular recognition of histone H3 by the WD40 protein WDR5.
Couture JF, Collazo E, Trievel RC. Couture JF, et al. Nat Struct Mol Biol. 2006 Aug;13(8):698-703. doi: 10.1038/nsmb1116. Epub 2006 Jul 9. Nat Struct Mol Biol. 2006. PMID: 16829960 - Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Ruthenburg AJ, et al. Nat Struct Mol Biol. 2006 Aug;13(8):704-12. doi: 10.1038/nsmb1119. Epub 2006 Jul 9. Nat Struct Mol Biol. 2006. PMID: 16829959 Free PMC article. - Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5.
Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J. Han Z, et al. Mol Cell. 2006 Apr 7;22(1):137-44. doi: 10.1016/j.molcel.2006.03.018. Mol Cell. 2006. PMID: 16600877 - On your histone mark, SET, methylate!
Binda O. Binda O. Epigenetics. 2013 May;8(5):457-63. doi: 10.4161/epi.24451. Epub 2013 Apr 27. Epigenetics. 2013. PMID: 23625014 Free PMC article. Review. - On WD40 proteins: propelling our knowledge of transcriptional control?
Migliori V, Mapelli M, Guccione E. Migliori V, et al. Epigenetics. 2012 Aug;7(8):815-22. doi: 10.4161/epi.21140. Epub 2012 Jul 19. Epigenetics. 2012. PMID: 22810296 Free PMC article. Review.
Cited by
- The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Zhang P, Lee H, Brunzelle JS, Couture JF. Zhang P, et al. Nucleic Acids Res. 2012 May;40(9):4237-46. doi: 10.1093/nar/gkr1235. Epub 2012 Jan 20. Nucleic Acids Res. 2012. PMID: 22266653 Free PMC article. - Epigenetic Reprogramming of the Glucose Metabolic Pathways by the Chromatin Effectors During Cancer.
Mondal P, Tiwary N, Sengupta A, Dhang S, Roy S, Das C. Mondal P, et al. Subcell Biochem. 2022;100:269-336. doi: 10.1007/978-3-031-07634-3_9. Subcell Biochem. 2022. PMID: 36301498 - Interaction of SET domains with histones and nucleic acid structures in active chromatin.
Krajewski WA, Vassiliev OL. Krajewski WA, et al. Clin Epigenetics. 2011 Apr;2(1):17-25. doi: 10.1007/s13148-010-0015-1. Epub 2011 Jan 14. Clin Epigenetics. 2011. PMID: 22704267 Free PMC article. - How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers.
Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. Taverna SD, et al. Nat Struct Mol Biol. 2007 Nov;14(11):1025-1040. doi: 10.1038/nsmb1338. Epub 2007 Nov 5. Nat Struct Mol Biol. 2007. PMID: 17984965 Free PMC article. Review. - Plant SET domain-containing proteins: structure, function and regulation.
Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC. Ng DW, et al. Biochim Biophys Acta. 2007 May-Jun;1769(5-6):316-29. doi: 10.1016/j.bbaexp.2007.04.003. Epub 2007 Apr 12. Biochim Biophys Acta. 2007. PMID: 17512990 Free PMC article. Review.
References
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D (2004) Regulation of p53 activity through lysine methylation. Nature 432: 353–360 - PubMed
- Couture JF, Collazo E, Hauk G, Trievel RC (2006a) Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol 13: 140–146 - PubMed
- Couture JF, Collazo E, Hauk G, Trievel RC (2006b) Molecular recognition of histone H3 by the WD40 protein. Nat Struct Mol Biol 13: 698–703 - PubMed
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 - PubMed
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases