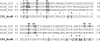The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons - PubMed (original) (raw)
The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons
Inna Grinberg et al. J Bacteriol. 2006 Nov.
Abstract
Ribonucleotide reductases (RNRs) catalyze the conversion of ribonucleotides to deoxyribonucleotides and are essential for de novo DNA synthesis and repair. Streptomyces spp. contain genes coding for two RNRs, either of which is sufficient for vegetative growth. The class Ia RNR is encoded by the nrdAB genes, and the class II RNR is encoded by nrdJ, which is coexpressed with nrdR. We previously showed that the Streptomyces coelicolor nrdR gene encodes a protein, NrdR, which represses transcription of both sets of RNR genes. NrdR is a member of a highly conserved family of proteins that is confined exclusively to prokaryotes. In this report, we describe a physical and biochemical characterization of the S. coelicolor NrdR protein and show that it is a zinc-ATP/dATP-containing protein that binds to the promoter regions of both Streptomyces RNR operons. The NrdR N terminus contains a zinc ribbon motif that is necessary for binding to the upstream regulatory region of both RNR operons. The latter contains two 16-bp direct repeat sequences, termed NrdR boxes, which are located proximal to, or overlap with, the promoter regions. These experiments support the view that NrdR controls the transcription of RNR genes by binding to the NrdR box sequences. We also show that the central NrdR ATP cone domain binds ATP and dATP and that mutations that abolish ATP/dATP binding significantly reduce DNA binding, suggesting that the ATP cone domain may allosterically regulate NrdR binding. We conclude that NrdR is a widely conserved regulator of RNR genes, binding to specific sequence elements in the promoter region and thereby modulating transcription.
Figures
FIG. 1.
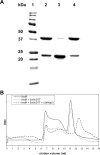
Molecular size and oligomeric properties of Ni-CAM affinity-purified recombinant NrdR. (A) SDS-PAGE results. Lanes: 1, protein molecular mass standards; 2, NrdR without any treatment; 3, NrdR heated to 100°C for 10 min; 4, NrdR with 50 mM DTT. (B) FPLC Superdex 200 gel filtration chromatography. Solid line, NrdR containing 0.3 M NaCl; dotted line, NrdR containing 5 mM DTT and 0.3 M NaCl; dashed line, NrdR containing 5 mM DTT and 1.5 M NaCl. Elution profiles monitored by the absorbance at 280 nm.
FIG. 2.
Multiple sequence alignment and domain organization of deduced NrdR proteins from representative members of major groups of bacteria. The proposed positions of the N-terminal zinc finger (residues 1 to 44) and the central ATP cone (residues 46 to 133) domains are indicated by overhead arrows. Conserved cysteine residues that define the zinc finger motif are highlighted (residues 3, 6, 31, and 34 by S. coelicolor numbering), as is a sequence of four consecutive arginine residues. Residues in the ATP cone domain presumed to be responsible for nucleotide binding are shown in Fig. 3. Symbols: asterisks, full conservation; colons, strongly similar; periods, weakly similar. Abbreviations: SCO, Streptomyces coelicolor (Actinobacteria); MTU, Mycobacterium tuberculosis (Actinobacteria); CGL, Corynebacterium glutamicum (Actinobacteria); BSU, Bacillus subtilis (Firmicutes, low GC content); MXA, Myxococcus xanthus (δ-Proteobacteria); TMA, Thermotoga maritima (Thermotogales); DRA, Deinococcus radiodurans; BPE, Bordetella pertussis (β-Proteobacteria); ATU, Agrobacterium tumefaciens (α-Proteobacteria); NPH, Natronomonas pharaonis (Euryarchaeota); ECO, Escherichia coli (γ-Proteobacteria).
FIG. 3.
Comparison of the amino acid sequence of the E. coli NrdA allosteric activity domain with that of the S. coelicolor and E. coli NrdR ATP cone domains to show conservation of ATP binding amino acid residues. NrdA residues reported to interact with an ATP analog (see text) are numbered; NrdA residues reported to interact with an ATP analog and which are conserved in S. coelicolor and E. coli NrdR are shown shaded. Two S. coelicolor NrdR residues that were mutated, Lys50 and Arg51, are shown boxed. The ATP cone domain consensus based on the deduced sequences of 250 NrdR proteins is shown at the bottom. Fully conserved NrdR residues are shown in bold and underlined. Symbols and abbreviations are as in the legend to Fig. 2. The function of the conserved sequence in the C-terminal region of the NrdR ATP cone domain, DEVAYVRFASVYRSF, is unknown.
FIG. 4.
UV absorption spectra of wild-type and mutant NrdR proteins. (A) Spectra of 0.04 mM solutions of the wild type and the Lys50Asn Arg51Gly double mutant. (B) Spectra of the supernatant obtained after perchloric acid treatment of 0.08 mM solutions of wild-type NrdR, NrdRΔNC, and NrdR Lys50Asn Arg51Gly double mutant. (C) Mono Q chromatography of NrdRΔNC. Peak fractions 1 and 2 did not contain protein and exhibited spectra matching that of adenine nucleotides (insert; see text for further details). (D) Affi-Gel boronate chromatography of the supernatant obtained after perchloric acid treatment of the NrdRΔNC mutant containing only the ATP cone domain. Standard solutions of dATP (left column), ATP (middle column), and the perchloric acid supernatant of NrdRΔNC (right column) were loaded onto an Affi-Gel column, and nucleotides were eluted, in succession, with buffer A to elute deoxyribonucleotides and with buffer B to release bound ribonucleotides. Amounts of released nucleotide were determined by HPLC and absorbance at 260 nm. OD, optical density.
FIG. 5.
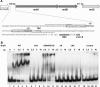
Binding of NrdR to the DNA region upstream of the S. coelicolor class Ia nrdABS RNR operon. (A) Nucleotide sequence of the DNA region containing the 84-bp probe used in binding assays. Primers used for preparing the PCR probe are indicated by dotted lines. The two NrdR boxes, shown by open rectangular frames, are located near to the transcription start site (TSS) and partially overlap with the promoter recognition element shown in bold italic. The putative ribosome binding site (RBS) and translation initiation codon GTG are shown in bold italic and underlined. (B) Binding of wild-type and mutant proteins to the nrdABS probe. DIG labeling of DNA probes and electrophoretic mobility shift assays were performed as described in Materials and Methods. Lanes: 1 to 5, NrdR wild type (WT) (0, 1, 2.5, 5, and 10 μg protein); 6 to 8, NrdR Cys3Ala (2.5, 5, and 10 μg protein); 9 to 11, NrdR Lys50Asn Arg51Gly (2.5, 5, and 10 μg protein); 12 to 14, NrdRΔN (2.5, 5, and 10 μg protein); 15 to 17, NrdRΔNC (2.5, 5, and 10 μg protein); 18 to 21, NrdR wild type (0, 2.5, 5, and 10 μg protein with the negative control cydA probe [84 bp]).
FIG. 6.
Binding of NrdR to the DNA region upstream of the S. coelicolor class II nrdRJ RNR operon. (A) Nucleotide sequence of the DNA region containing the 84-bp probe used in binding assays. Primers used for preparing the PCR probe are indicated by dotted lines. The two NrdR boxes are shown by open rectangular frames according to the work of Rodionov and Gelfand (21), and our prediction of NrdR binding sites is indicated with solid arrows above the nucleotides shown in bold. The putative ribosome binding site (RBS) and translation initiation codon ATG are shown in bold italic and underlined. (B) Binding of wild-type and mutant proteins to the nrdRJ probe. DIG labeling of DNA probes and electrophoretic mobility shift assays were performed as described in Materials and Methods. Lanes: 1 to 5, NrdR wild type (WT) (0, 1, 2.5, 5, and 10 μg protein); 6 to 8, NrdR Cys3Ala (2.5, 5, and 10 μg protein); 9 to 11, NrdR Lys50Asn Arg51Gly (2.5, 5, and 10 μg protein); 12 and 13, NrdRΔN (5 and 10 μg protein); 14 and 15, NrdRΔNC (5 and 10 μg protein); 16 to 18, NrdRΔC (2.5, 5, and 10 μg protein).
Similar articles
- Functional analysis of the Streptomyces coelicolor NrdR ATP-cone domain: role in nucleotide binding, oligomerization, and DNA interactions.
Grinberg I, Shteinberg T, Hassan AQ, Aharonowitz Y, Borovok I, Cohen G. Grinberg I, et al. J Bacteriol. 2009 Feb;191(4):1169-79. doi: 10.1128/JB.01145-08. Epub 2008 Dec 1. J Bacteriol. 2009. PMID: 19047342 Free PMC article. - Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation.
Borovok I, Gorovitz B, Yanku M, Schreiber R, Gust B, Chater K, Aharonowitz Y, Cohen G. Borovok I, et al. Mol Microbiol. 2004 Nov;54(4):1022-35. doi: 10.1111/j.1365-2958.2004.04325.x. Mol Microbiol. 2004. PMID: 15522084 - NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes.
Torrents E, Grinberg I, Gorovitz-Harris B, Lundström H, Borovok I, Aharonowitz Y, Sjöberg BM, Cohen G. Torrents E, et al. J Bacteriol. 2007 Jul;189(14):5012-21. doi: 10.1128/JB.00440-07. Epub 2007 May 11. J Bacteriol. 2007. PMID: 17496099 Free PMC article. - Structures and organisation of AAA+ enhancer binding proteins in transcriptional activation.
Schumacher J, Joly N, Rappas M, Zhang X, Buck M. Schumacher J, et al. J Struct Biol. 2006 Oct;156(1):190-9. doi: 10.1016/j.jsb.2006.01.006. Epub 2006 Feb 20. J Struct Biol. 2006. PMID: 16531068 Review. - Spectroscopic and theoretical approaches for studying radical reactions in class I ribonucleotide reductase.
Bennati M, Lendzian F, Schmittel M, Zipse H. Bennati M, et al. Biol Chem. 2005 Oct;386(10):1007-22. doi: 10.1515/BC.2005.117. Biol Chem. 2005. PMID: 16218873 Review.
Cited by
- The tRNA thiolation pathway modulates the intracellular redox state in Escherichia coli.
Nakayashiki T, Saito N, Takeuchi R, Kadokura H, Nakahigashi K, Wanner BL, Mori H. Nakayashiki T, et al. J Bacteriol. 2013 May;195(9):2039-49. doi: 10.1128/JB.02180-12. Epub 2013 Mar 1. J Bacteriol. 2013. PMID: 23457245 Free PMC article. - An endogenous dAMP ligand in Bacillus subtilis class Ib RNR promotes assembly of a noncanonical dimer for regulation by dATP.
Parker MJ, Maggiolo AO, Thomas WC, Kim A, Meisburger SP, Ando N, Boal AK, Stubbe J. Parker MJ, et al. Proc Natl Acad Sci U S A. 2018 May 15;115(20):E4594-E4603. doi: 10.1073/pnas.1800356115. Epub 2018 Apr 30. Proc Natl Acad Sci U S A. 2018. PMID: 29712847 Free PMC article. - NrdR Transcription Regulation: Global Proteome Analysis and Its Role in Escherichia coli Viability and Virulence.
Naveen V, Hsiao CD. Naveen V, et al. PLoS One. 2016 Jun 8;11(6):e0157165. doi: 10.1371/journal.pone.0157165. eCollection 2016. PLoS One. 2016. PMID: 27275780 Free PMC article. - Insights from the architecture of the bacterial transcription apparatus.
Iyer LM, Aravind L. Iyer LM, et al. J Struct Biol. 2012 Sep;179(3):299-319. doi: 10.1016/j.jsb.2011.12.013. Epub 2011 Dec 24. J Struct Biol. 2012. PMID: 22210308 Free PMC article. - Functional analysis of the Streptomyces coelicolor NrdR ATP-cone domain: role in nucleotide binding, oligomerization, and DNA interactions.
Grinberg I, Shteinberg T, Hassan AQ, Aharonowitz Y, Borovok I, Cohen G. Grinberg I, et al. J Bacteriol. 2009 Feb;191(4):1169-79. doi: 10.1128/JB.01145-08. Epub 2008 Dec 1. J Bacteriol. 2009. PMID: 19047342 Free PMC article.
References
- Aravind, L., Y. I. Wolf, and E. V. Koonin. 2000. The ATP-cone: an evolutionarily mobile, ATP-binding regulatory domain. J. Mol. Microbiol. Biotechnol. 2:191-194. - PubMed
- Borovok, I., B. Gorovitz, M. Yanku, R. Schreiber, B. Gust, K. Chater, Y. Aharonowitz, and G. Cohen. 2004. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 54:1022-1035. - PubMed
- Borovok, I., R. Kreisberg-Zakarin, M. Yanko, R. Schreiber, M. Myslovati, F. Aslund, A. Holmgren, G. Cohen, and Y. Aharonowitz. 2002. Streptomyces spp. contain class Ia and class II ribonucleotide reductases: expression analysis of the genes in vegetative growth. Microbiology 148:391-404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous