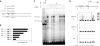Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells - PubMed (original) (raw)
Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells
Yuhki Nakatake et al. Mol Cell Biol. 2006 Oct.
Abstract
Although the POU transcription factor Oct3/4 is pivotal in maintaining self renewal of embryonic stem (ES) cells, little is known of its molecular mechanisms. We previously reported that the N-terminal transactivation domain of Oct3/4 is required for activation of Lefty1 expression (H. Niwa, S. Masui, I. Chambers, A. G. Smith, and J. Miyazaki, Mol. Cell. Biol. 22:1526-1536, 2002). Here we test whether Lefty1 is a direct target of Oct3/4. We identified an ES cell-specific enhancer upstream of the Lefty1 promoter that contains binding sites for Oct3/4 and Sox2. Unlike other known Oct3/4-Sox2-dependent enhancers, however, this enhancer element could not be activated by Oct3/4 and Sox2 in differentiated cells. By functional screening of ES-specific transcription factors, we found that Krüppel-like factor 4 (Klf4) cooperates with Oct3/4 and Sox2 to activate Lefty1 expression, and that Klf4 acts as a mediating factor that specifically binds to the proximal element of the Lefty1 promoter. DNA microarray analysis revealed that a subset of putative Oct3/4 target genes may be regulated in the same manner. Our findings shed light on a novel function of Oct3/4 in ES cells.
Figures
FIG. 1.
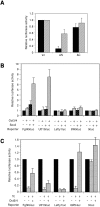
Identification of an Oct3/4-dependent enhancer in the Lefty1 gene. (A) Isolation of the mouse Lefty1 promoter. A 1,379-bp genomic DNA fragment, from −1297 to +82 relative to the transcription start site, was subcloned by PCR. Ch. 1, chromosome 1. (B) Oct3/4-dependent promoter activity of pLefty1-luc in ES cells. Reporter plasmids were transfected into ZHBTc4 ES cells, followed by culture with or without Tc. The ratios (−Tc/+Tc) of the resulting luciferase activities are shown. (C and D) Oct3/4-dependent activities of deletion mutants of pLefty1-luc. (E and F) Enhancer activity of the element containing the putative Oct-Sox binding site. The activities of small deletion mutants (E) and pLefty1-luc with mutations in the Oct or Sox element (F) were assayed in ZHBTc4 ES cells (F). (G) EMSA of the Oct-Sox element. Wild-type (wt) Oct-Sox elements were incubated with ES nuclear extracts, with or without excess cold competitors (lanes 1 to 7), or incubated with antibodies to Oct3/4, Sox2, or Gata4 (lanes 8 to 10), as indicated, and subjected to EMSA. The sequences of the probes are shown in Table 1. Note that the supershifted bands (ss) and reduction of signal were obtained with each antibody.
FIG. 2.
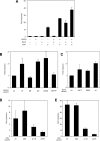
Activation of Lefty1 promoter by Oct3/4 and Sox2. (A) Activities of pLefty1-luc in ES cells maintained by mutant forms of Oct3/4. Relative expression levels of plefty1-luc (filled) and Fgf4tkluc (hatched) are shown. Fgf4tkluc has a tk minimal promoter ligated with the enhancer elements in which Oct3/4 and Sox2 can associate. The luciferase activity in ES cells expressing wild-type (wt) Oct3/4 was set at 1.0. (B) Relative activation of reporters by Oct3/4 and Sox2 in HeLa cells. The reporter plasmids were cotransfected with Oct3/4 and/or Sox2 expression vectors. The luciferase activity of reporter alone was set at 1.0. (C) Reactivation of reporters by Oct3/4 in _Oct3/4_-depleted ES cells. Reporter plasmids were transfected into ZHBTc4 ES cells, with or without Oct3/4 expression vector, and the cells were cultured with or without Tc. Resulting luciferase activities, relative to that in ES cells without Tc and Oct3/4 expression vector, are shown.
FIG. 3.
Activation of the Lefty1 reporter by Klf4. (A) Activation of pLefty1-luc by various ES-specific transcription factors with (open) or without (filled) Oct3/4 and Sox2 expression vectors in HeLa cells. Induction of luciferase activity was measured relative to that of empty vector. (B) Activation of various reporters by Klf4 in HeLa cells. Reporter plasmids were cotransfected with empty or Klf4 expression vector into HeLa cells and the luciferase activities, relative to that of empty vector, were measured. (C) Activation of pLefty1-luc by Klf family members. pLefty1-luc was cotransfected with empty or Klf family expression vectors into HeLa cells and luciferase activities, relative to that of empty vector, were measured.
FIG. 4.
Cooperation of Oct3/4, Sox2, and Klf4 in activating the Lefty1 reporter. (A) Cooperative activation of the Lefty1 reporter by Oct3/4, Sox2, and Klf4. pLefty1-luc was cotransfected with various combinations of empty, Oct3/4, Sox2, and Klf4 expression vectors into HeLa cells, and luciferase activities were measured relative to that of empty vector. (B) Activation of the Lefty1 reporter by Oct3/4 variants. pLefty1-luc was cotransfected with expression vectors of Oct3/4 variants, Sox2, and Klf4 into HeLa cells, and luciferase activities were measured relative to that of empty vector. (C) Activation of the Lefty1 reporter by Sox2 variants. pLefty1-luc was cotransfected with expression vectors of Sox2 variants, Oct3/4, and Klf4 into HeLa cells, and luciferase activities were measured relative to that of empty vector. (D and E) Activation of the Lefty1 reporter by Klf4 variants. pLefty1-luc was cotransfected with expression vectors of Klf4 variants alone (D) or Klf4 variants plus Oct3/4 and Sox2 (E) into HeLa cells, and luciferase activities were measured relative to that of empty vector. wt, wild type.
FIG. 5.
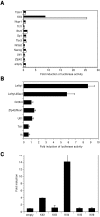
Identification of Klf4 binding sites proximal to the Lefty1 core promoter. (A) Sequence around the Lefty1 core promoter in pLefty1ΔSacI-luc. Two putative Klf4 binding sites (K1 and K2) were identified proximal to the core promoter, both of which are conserved in human LEFTYB. Mutated sequences for K1m and K2m are also shown. (B) Contribution of the putative Klf4 binding sites to the Klf4-dependent activity of pLefty1ΔSacI-luc. pLefty1ΔSacI-luc derivatives with mutations in K1 and/or K2 or deletion of both sites were cotransfected with Klf4 or empty expression vector into HeLa cells, and luciferase activities were determined relative to that of empty vector. (C) Cell lysate of ES or Cos-7 cells transfected with Klf4 or empty expression vector was subjected to EMSA with or without competitor. All probes are described in Table 2. (D) ChIP assay of the Lefty1 distal enhancer, proximal element, and core promoter. Primers for QPCR were designed to detect the distal enhancer bound by Oct3/4 and Sox2 (region b), the proximal element bound by Klf4 and the core promoter (region d), and their flanking regions (a, c, and e) as controls. Chromatin samples derived from undifferentiated (ZHBTc4 Tc−), differentiated (ZHBTc4 Tc+), and _Δ_N ES cells were immunoprecipitated with the indicated antibody, followed by QPCR with normalization by input DNA. The amounts of region a were set at 1.0 in each set of analyses, and the relative signal intensities for regions b, c, d, and e were calculated.
FIG. 6.
Klf4 target genes in ES cells. (A) Expression of stem cell marker genes in ES cells treated with siKlf4. Relative expression levels of each gene in ES cells treated with mock transfectant (filled; set at 1.0) or siKlf4 (hatched) was estimated by QPCR. (B) DNA microarray analyses of ES cells treated with siKlf4. Log ratios were plotted for genes with relative expression levels. The plot for Lefty1 is indicated. (C) Identification of putative target genes regulated by both Klf4 and Oct3/4. We identified 28 genes whose expression was dependent on both Klf4 and Oct3/4. (D) Activation of the 1200015N20Rik promoter by Klf4. 1200015N20Rik-luc, 6Wtk-luc, or tk-luc was cotransfected with empty or Klf4 expression vector into HeLa cells, and the luciferase activities relative to empty vector were determined. (E) Oct3/4 reactivation of reporters in _Oct3/4_-depleted ES cells. Reporter plasmids were transfected into ZHBTc4 ES cells with or without Oct3/4 expression vector, the cells were cultured with or without Tc, and luciferase activities relative to that in ES cells without Tc and Oct3/4 expression vector were determined.
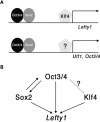
FIG. 7.
Role of Klf4 in the activation of the Lefty1 promoter. (A) Differential promoter factor requirement for activation by Oct3/4 and Sox2. The Lefty1 promoter is activated by recruitment of Klf4 as a promoter factor, but requirements for the Utf1 and Oct3/4 promoters have not yet been identified. (B) Regulation of Lefty1 by ES-specific transcription factors. Oct3/4 and Sox2 are activated by themselves, and Klf4 is under the control of Oct3/4.
Similar articles
- Zfp296 is a novel Klf4-interacting protein and functions as a negative regulator.
Fujii Y, Kakegawa M, Koide H, Akagi T, Yokota T. Fujii Y, et al. Biochem Biophys Res Commun. 2013 Nov 15;441(2):411-7. doi: 10.1016/j.bbrc.2013.10.073. Epub 2013 Oct 22. Biochem Biophys Res Commun. 2013. PMID: 24161396 - Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells.
Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Masui S, et al. Nat Cell Biol. 2007 Jun;9(6):625-35. doi: 10.1038/ncb1589. Epub 2007 May 21. Nat Cell Biol. 2007. PMID: 17515932 - Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells.
Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Maruyama M, et al. J Biol Chem. 2005 Jul 1;280(26):24371-9. doi: 10.1074/jbc.M501423200. Epub 2005 Apr 29. J Biol Chem. 2005. PMID: 15863505 - Conserved and divergent paths that regulate self-renewal in mouse and human embryonic stem cells.
Rao M. Rao M. Dev Biol. 2004 Nov 15;275(2):269-86. doi: 10.1016/j.ydbio.2004.08.013. Dev Biol. 2004. PMID: 15501218 Review. - Mouse induced pluripotent stem cells.
Geoghegan E, Byrnes L. Geoghegan E, et al. Int J Dev Biol. 2008;52(8):1015-22. doi: 10.1387/ijdb.082640eg. Int J Dev Biol. 2008. PMID: 18956334 Review.
Cited by
- Cancer Stem Cell Hierarchy in Glioblastoma Multiforme.
Bradshaw A, Wickremsekera A, Tan ST, Peng L, Davis PF, Itinteang T. Bradshaw A, et al. Front Surg. 2016 Apr 15;3:21. doi: 10.3389/fsurg.2016.00021. eCollection 2016. Front Surg. 2016. PMID: 27148537 Free PMC article. Review. - Emerging stem cell therapies: treatment, safety, and biology.
Sng J, Lufkin T. Sng J, et al. Stem Cells Int. 2012;2012:521343. doi: 10.1155/2012/521343. Epub 2012 Aug 2. Stem Cells Int. 2012. PMID: 22919402 Free PMC article. - Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming.
Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Soufi A, et al. Cell. 2015 Apr 23;161(3):555-568. doi: 10.1016/j.cell.2015.03.017. Epub 2015 Apr 16. Cell. 2015. PMID: 25892221 Free PMC article. - Rapid activation of the bivalent gene Sox21 requires displacement of multiple layers of gene-silencing machinery.
Chakravarthy H, Ormsbee BD, Mallanna SK, Rizzino A. Chakravarthy H, et al. FASEB J. 2011 Jan;25(1):206-18. doi: 10.1096/fj.10-166926. Epub 2010 Sep 27. FASEB J. 2011. PMID: 20876214 Free PMC article. - The miR-26a/AP-2α/Nanog signaling axis mediates stem cell self-renewal and temozolomide resistance in glioma.
Huang W, Zhong Z, Luo C, Xiao Y, Li L, Zhang X, Yang L, Xiao K, Ning Y, Chen L, Liu Q, Hu X, Zhang J, Ding X, Xiang S. Huang W, et al. Theranostics. 2019 Jul 28;9(19):5497-5516. doi: 10.7150/thno.33800. eCollection 2019. Theranostics. 2019. PMID: 31534499 Free PMC article.
References
- Aiba, K., A. A. Sharov, M. G. Carter, C. Foroni, A. L. Vescovi, and M. S. Ko. 2006. Defining a developmental path to neural fate by global expression profiling of mouse embryonic stem cells and adult neural stem/progenitor cells. Stem Cells 24:889-895. - PubMed
- Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. - PMC - PubMed
- Bertolino, E., and H. Singh. 2002. POU/TBP cooperativity: a mechanism for enhancer action from a distance. Mol. Cell. 10:397-407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials