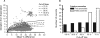New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras - PubMed (original) (raw)
Comparative Study
New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras
Kevin E Ashelford et al. Appl Environ Microbiol. 2006 Sep.
Abstract
A new computer program, called Mallard, is presented for screening entire 16S rRNA gene libraries of up to 1,000 sequences for chimeras and other artifacts. Written in the Java computer language and capable of running on all major operating systems, the program provides a novel graphical approach for visualizing phylogenetic relationships among 16S rRNA gene sequences. To illustrate its use, we analyzed most of the large libraries of cloned bacterial 16S rRNA gene sequences submitted to the public repository during 2005. Defining a large library as one containing 100 or more sequences of 1,200 bases or greater, we screened 25 of the 28 libraries and found that all but three contained substantial anomalies. Overall, 543 anomalous sequences were found. The average anomaly content per clone library was 9.0%, 4% higher than that previously estimated for the public repository overall. In addition, 90.8% of anomalies had characteristic chimeric patterns, a rise of 25.4% over that found previously. One library alone was found to contain 54 chimeras, representing 45.8% of its content. These figures far exceed previous estimates of artifacts within public repositories and further highlight the urgent need for all researchers to adequately screen their libraries prior to submission. Mallard is freely available from our website at http://www.cardiff.ac.uk/biosi/research/biosoft/.
Figures
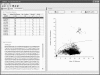
FIG. 1.
Mallard program screenshot, illustrating a typical analysis. In this example, the library containing 222 16S rRNA gene sequences representing the Verrucomicrobia phylum is being considered. Each sequence within the library was compared with every other sequence, generating 24,531 separate DE values that were plotted against the mean percentage differences (a simple measure of evolutionary distance). Unusually high DE values are those plotted above the superimposed dotted line, and they represent comparisons in which one (or both) of the sequences is likely to be anomalous. From these outlier DE values, a list of suspected anomalies is generated (upper left-hand panel of the screenshot). Clicking on a listed sequence record causes associated DE values to be highlighted in red in the right-hand panel. Clicking on individual plotted DE values displays the underlying Pintail plot in a separate panel (not shown), and from this information, the nature of any anomaly may be discerned.
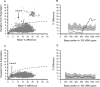
FIG. 2.
Mallard-generated DE plot in detail. (A) Reproduced DE plot of the Verrucomicrobia phylum library shown in Fig. 1 with the dotted line (the 100% cutoff line) identifying unusually high DE values (outliers), which lie above the line. Each plotted DE value represents a separate sequence comparison using the Pintail algorithm, and clicking on a plotted point within the program reveals the underlying Pintail plot. (B) The plot generated from one such comparison (between the chimera AY752110 and the error-free AF050561). The solid black line represents changes in evolutionary distance between these two sequences, when aligned, as determined from a 300-base sampling window moving 25 bases at a time along the alignment (2). The solid dark-gray line represents those evolutionary distances that one might have expected had both sequences been error free (2). The disparity between these two lines reflects the chimeric nature of AY752110. Excluding this and other chimeras identified by the program from the analysis produces the plot in panel C. DE values below the dotted cutoff line result from comparisons between error-free sequences; panel D represents a typical example, with AY212657 being compared with AB154319.
FIG. 3.
Impact of cutoff line choice on correct identification of anomalies. (A) DE values from the phylum Verrucomicrobia analysis are plotted, with the five possible cutoff lines superimposed. (B) The numbers of true anomalies and false positives recorded for each cutoff line show that reducing the cutoff line allows more actual anomalies to be correctly identified as such but also leads to an increased number of falsely identified anomalies. The default cutoff line for the Mallard program is 99.9%, which provides a reasonable compromise between detecting as many anomalies as possible and producing the smallest number of false positives.
FIG. 4.
Analysis of near-complete (≥1,200-base) sequences from 25 16S rRNA gene clone libraries submitted to the public repositories during 2005 (5-12, 15, 18, 21, 25, 28, 32, 33). Gene libraries are identified by the first author surname and the RDP REFID number, with the number of near-complete sequences (library size) in parentheses. The bars indicate the number of detected anomalies (identified with the 100% cutoff line) as a percentage of library size, with those anomalies confirmed as such by further investigation and false positives shown.
References
- Cole, J., B. Chai, T. Marsh, R. Farris, Q. Wang, S. Kulum, S. Chandra, D. McGarrell, T. Schmidt, G. Garrity, and J. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. - PMC - PubMed
- Crump, B. C., and J. E. Hobbie. 2005. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol. Oceanogr. 50:1718-1729.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources