The bacterial cytoskeleton - PubMed (original) (raw)
Review
The bacterial cytoskeleton
Yu-Ling Shih et al. Microbiol Mol Biol Rev. 2006 Sep.
Abstract
In recent years it has been shown that bacteria contain a number of cytoskeletal structures. The bacterial cytoplasmic elements include homologs of the three major types of eukaryotic cytoskeletal proteins (actin, tubulin, and intermediate filament proteins) and a fourth group, the MinD-ParA group, that appears to be unique to bacteria. The cytoskeletal structures play important roles in cell division, cell polarity, cell shape regulation, plasmid partition, and other functions. The proteins self-assemble into filamentous structures in vitro and form intracellular ordered structures in vivo. In addition, there are a number of filamentous bacterial elements that may turn out to be cytoskeletal in nature. This review attempts to summarize and integrate the in vivo and in vitro aspects of these systems and to evaluate the probable future directions of this active research field.
Figures
FIG. 1.
Structural comparison of eukaryotic and prokaryotic cytoskeletal proteins. Protein structures were downloaded from the Protein Data Bank (PDB) (
) and aligned using the EMBL-EBI Dali server (
). Protein structures were then analyzed and displayed using the MolMol program (103). Nucleotides and inorganic molecules that cocrystalized with the proteins are not shown. (A) Actin homologs. From left to right: Saccharomyces cerevisiae actin (PDB entry 1YAGA), ATP bound (218); Thermotoga maritima MreB (PDB entry 1JCG), AMPPNP bound (214); ParM of plasmid R1 (PDB entry 1MWM), ADP bound (216). (B) Tubulin homologs. From left to right: Bos taurus β-tubulin (PDB entry 1JFFB), GTP/GDP bound (124); Methanococcus jannaschii FtsZ (PDB entry 1FSZ), GDP bound (122); Prosthecobacter dejongeii BtubA (PDB entry 2BTOA), GTP bound (183). (C) MinD (left) and Soj (right) proteins: Archaeoglobus fulgidus MinD (PDB entry 1HYQ), nucleotide free (24); Thermus thermophilus Soj (PDB entry 2BEJ), ADP bound (116). The topological specificity domain of the MinE dimer is shown (residues 31 to 88). The MinD-binding domains of each monomer extend from opposite sides (the right and left sides in this view) of the dimeric topological specificity domain (100).
FIG. 2.
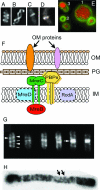
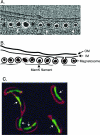
Localization and functions of the actin-like MreB protein. (A to D) Cellular localization of MreB labeled with GFP or YFP. (A to C) E. coli MreB localizes into extended coils (A), intertwined double helices (B), and band-like structures (C). (Reprinted from reference with permission of the publisher. Copyright 2003 National Academy of Sciences, U.S.A.) (D) C. crescentus MreB localizes into a band-like structure at the division site in predivisional cells. (Reprinted from reference with permission of the publisher. Copyright 2004 National Academy of Sciences, U.S.A.) (E) Chromosome segregation in E. coli mreB deletion strain (Y.-L. Shih, unpublished). Defects include multiple disordered chromosomes and unequal chromosome partition into daughter cells (cell 1), production of anucleate cells (cell 2), and chromosome fragmentation presumably due to guillotining of the nucleoid (cell 3). Bacterial DNA (red) is stained with DAPI; membrane (green) is stained with FM4-64 [_N_-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide]. (F) Suggested organization of cytoskeleton-associated cell wall-synthesizing proteins of E. coli. The cytoplasmic MreB cytoskeleton is linked via MreC and MreD to the PBP murein biosynthetic enzymes (107). RodA may also be a member of the complex, and it is possible that some outer membrane proteins are also part of the MreC-associated structure (35). MreC is shown as a transmembrane protein (107, 113), but, at least in C. crescentus, MreC appears to be a periplasmic protein (35). See text for further details and references. This multiprotein structure may permit the MreB cytoskeleton to regulate the pattern of cell wall biosynthesis by providing spatial information to the murein biosynthetic machinery. OM, outer membrane; PG, peptidoglycan (murein); IM, inner membrane. (G) Pattern of murein assembly in B. subtilis (27). Nascent peptidoglycan was stained with fluorescently labeled vancomycin (see the text). The micrograph contains cells representing different stages of the cell cycle. Helical patterns of labeled vancomycin are indicated by transverse bands (lines) and dots around the cell periphery (arrowheads). The densely stained bands (arrow) in the two right-most cells are located at the cell division sites. (Reprinted from reference with permission from Elsevier.) (H) Pattern of murein insertion into the murein sacculus of E. coli, suggesting a helical mode of murein assembly (arrows indicate two of the helical coils). (Reprinted from reference with permission of the publisher.)
FIG. 3.
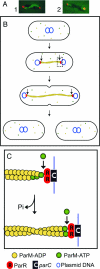
ParM-mediated partition of plasmid R1. (A) Fluorescence micrographs showing the ParM filament (green) and plasmid DNA (red). Panel 1 shows a ParM filament extending between two plasmids that are located at the ends of the cell. Panel 2 shows a plasmid at each cell pole, attached to a short ParM filament that extends toward midcell; this presumably represents an intermediate stage of filament disassembly after the plasmids have been delivered to the ends of the cell. (Reprinted from reference with permission from Elsevier.) (B) After plasmid replication, growth of a ParM polymeric filament (yellow) pushes apart the two progeny R1 plasmids, moving them from midcell to the two poles. (C) Cartoon representing the boxed region in panel B, showing details of the addition of new subunits at the two ends of the ParM filament. The ParM-ADP filament is capped by ParM-ATP subunits. The capped end is attached to the parC centromeric region of the plasmid via the ParR protein. ParM filament growth occurs by insertion of ParM-ATP subunits at the interface between the end of the ParM filament and the ParR/parC complex. This is associated with hydrolysis of the ATP moiety of the previous ParM-ATP subunit, thereby converting the penultimate subunit to ParM-ADP.
FIG. 4.
Intracellular cytoskeletal structures formed by the actin-like MamK protein (A and B) and the intermediate filament homolog crescentin (C). (A and B) Electron cryotomography (A) and cartoon illustration (B) of magnetosomes located under the surface of the cytoplasmic membrane of Magnetospirillum magneticum. MamK filaments extend along the surface of the linear array of membrane-bounded magnetosome structures (white arrows in panel A). (The electron micrograph was reprinted from reference with permission of AAAS.) (C) The intermediate filament-like protein crescentin (green), visualized by expressing GFP-tagged crescentin in wild-type cells, forms filamentous structures (white arrows) along the concave edge of C. crescentus. Membranes (in red) are stained with dye FM 4-64. (Reprinted from reference with permission from Elsevier.)
FIG. 5.
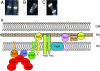
Cellular organization of FtsZ. (A to C) Immunofluorescence micrographs showing cellular localization of FtsZ in B. subtilis cells. (Reprinted from reference with permission from Elsevier.) (A) FtsZ ring at midcell during vegetative growth; (B) extended FtsZ helical structures during the transition from vegetative growth to sporulation; (C) bipolar FtsZ rings at a later stage of the transition. (D) Diagrammatic representation of a portion of the cytokinetic ring (septasome) of E. coli. FtsZ is anchored to the cell membrane by FtsA and ZipA, and the other septasomal proteins are then added to the complex (63). The symbols representing the proteins are arranged for clarity, and the cartoon does not accurately represent the details of their interactions. OM, outer membrane; PG, peptidoglycan (murein); IM, inner membrane.
FIG. 6.
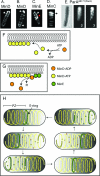
Helical organization of the MinD/ParA cytoskeletal proteins. (A to D) Fluorescently labeled MinD (A and B), MinE (C), and MinC (D). (Reprinted from reference with permission of the publisher. Copyright 2003 National Academy of Sciences, U.S.A.) (A and B) Helical distribution of MinD when coexpressed with MinE. (A) A bright MinD helical structure is present at one end of the cell, representing a MinD polar zone (white arrow). A less intense coiled structure extends to the other end of the cell, presumably representing the underlying pole-to-pole MinD helical structure. (B) Short MinD polar zones are visible at both ends of the cell (white arrows), one presumably representing a growing zone and the other representing a polar zone at the opposite end of the cell, in the last stage of disassembly, as depicted in panel H. (C) Helical distribution of MinE when coexpressed with MinD. The majority of the MinE molecules are present in an end loop (the MinE ring, red arrow) of the MinE helical array that represents the polar zone. The white arrow indicates polar zone coils. Micrographs in panels A to C are deconvolved images. (D) Helical distribution of MinC when coexpressed with MinD and MinE (three-dimensional reconstruction from an optically sectioned cell). The arrow indicates the MinC coils of the polar zone. (E) Helical distribution of fluorescently labeled ParA from plasmid pB171. 1, Nomarski image; 2, raw image; 3, deconvolved image. (Reprinted from reference with permission from Blackwell Publishing.) (F) Mechanism of MinD polymer growth on the cytoplasmic face of the inner membrane by addition of MinD-ATP subunits. (G) Stimulation of disassembly of MinD-ATP polymer by a MinE ring. MinE molecules in the E-ring stimulate the ATPase activity of the terminal MinD-ATP subunit in the polymer. Hydrolysis converts MinD-ATP to MinD-ADP, which is released from the end of the polymer. (H) Cyclic assembly and disassembly of the MinD helical polar zones (PZ) and MinE ring, leading to pole-to-pole oscillations. MinD-ATP subunits are yellow, MinD-ADP subunits are orange, and MinE subunits are green. See text for details. The solid yellow helical structures represent the underlying MinD helical structure that extends between the two ends of the cell. MinD helical structures also contain MinE and MinC, which are not shown for simplification.
FIG. 7.
In vitro polymerization of cytoskeletal proteins of the MinD/ParA superfamily. (A) Formation of MinD filament bundles in the presence of MinE, ATP, and phospholipid vesicles. One end of the bundle is markedly frayed because of the presence of MinE. (Reprinted from reference with permission of the publisher. Copyright 2003 National Academy of Sciences, U.S.A.) (B) Formation of a ParApTP228(ParF) filament bundle in the presence of ParBpTP228(ParG) and ATP. ParBpTP228(ParG) stimulates formation of the frayed end(s) of the ParApTP228(ParF) bundle. (Reprinted from reference by permission from Macmillan Publishers Ltd.) (C) Formation of Soj filaments in the presence of DNA and ATP. (Reprinted from reference by permission from Macmillan Publishers Ltd.)
Similar articles
- Dynamics of bacterial cytoskeletal elements.
Graumann PL. Graumann PL. Cell Motil Cytoskeleton. 2009 Nov;66(11):909-14. doi: 10.1002/cm.20381. Cell Motil Cytoskeleton. 2009. PMID: 19466751 Review. - Dynamic filaments of the bacterial cytoskeleton.
Michie KA, Löwe J. Michie KA, et al. Annu Rev Biochem. 2006;75:467-92. doi: 10.1146/annurev.biochem.75.103004.142452. Annu Rev Biochem. 2006. PMID: 16756499 Review. - Bacterial Filament Systems: Toward Understanding Their Emergent Behavior and Cellular Functions.
Eun YJ, Kapoor M, Hussain S, Garner EC. Eun YJ, et al. J Biol Chem. 2015 Jul 10;290(28):17181-9. doi: 10.1074/jbc.R115.637876. Epub 2015 May 8. J Biol Chem. 2015. PMID: 25957405 Free PMC article. Review. - Cytoskeletal elements in bacteria.
Graumann PL. Graumann PL. Annu Rev Microbiol. 2007;61:589-618. doi: 10.1146/annurev.micro.61.080706.093236. Annu Rev Microbiol. 2007. PMID: 17506674 Review. - The role of cytoskeletal elements in shaping bacterial cells.
Cho H. Cho H. J Microbiol Biotechnol. 2015 Mar;25(3):307-16. doi: 10.4014/jmb.1409.09047. J Microbiol Biotechnol. 2015. PMID: 25262683 Review.
Cited by
- Plasmid segregation without partition.
Guynet C, de la Cruz F. Guynet C, et al. Mob Genet Elements. 2011 Sep;1(3):236-241. doi: 10.4161/mge.1.3.18229. Epub 2011 Sep 1. Mob Genet Elements. 2011. PMID: 22312593 Free PMC article. - DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum.
Letek M, Ordóñez E, Vaquera J, Margolin W, Flärdh K, Mateos LM, Gil JA. Letek M, et al. J Bacteriol. 2008 May;190(9):3283-92. doi: 10.1128/JB.01934-07. Epub 2008 Feb 22. J Bacteriol. 2008. PMID: 18296522 Free PMC article. - An Expanded Transposon Mutant Library Reveals that Vibrio fischeri δ-Aminolevulinate Auxotrophs Can Colonize Euprymna scolopes.
Lyell NL, Septer AN, Dunn AK, Duckett D, Stoudenmire JL, Stabb EV. Lyell NL, et al. Appl Environ Microbiol. 2017 Feb 15;83(5):e02470-16. doi: 10.1128/AEM.02470-16. Print 2017 Mar 1. Appl Environ Microbiol. 2017. PMID: 28003196 Free PMC article. - Label-Free Proteomics of a Defined, Binary Co-culture Reveals Diversity of Competitive Responses Between Members of a Model Soil Microbial System.
Chignell JF, Park S, Lacerda CMR, De Long SK, Reardon KF. Chignell JF, et al. Microb Ecol. 2018 Apr;75(3):701-719. doi: 10.1007/s00248-017-1072-1. Epub 2017 Oct 3. Microb Ecol. 2018. PMID: 28975425
References
- Adachi, S., K. Hori, and S. Hiraga. 2006. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J. Mol. Biol. 356:850-863. - PubMed
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular biology of the cell, 4th ed. Garland Science, New York, N.Y.
- Aldridge, C., J. Maple, and S. Moller. 2005. The molecular biology of plastid division in higher plants. J. Exp. Bot. 56:1061-1077. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources