A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping - PubMed (original) (raw)
A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping
Chao Tian et al. Am J Hum Genet. 2006 Oct.
Abstract
Admixture mapping requires a genomewide panel of relatively evenly spaced markers that can distinguish the ancestral origins of chromosomal segments in admixed individuals. Through use of the results of the International HapMap Project and specific selection criteria, the current study has examined the ability of selected single-nucleotide polymorphisms (SNPs) to extract continental ancestry information in African American subjects and to explore parameters for admixture mapping. Genotyping of two linguistically diverse West African populations (Bini and Kanuri Nigerians, who are Niger-Congo [Bantu] and Nilo-Saharan speakers, respectively), European Americans, and African Americans validated a genomewide set of >4,000 SNP ancestry-informative markers with mean and median F(ST) values >0.59 and mean and median Fisher's information content >2.5. This set of SNPs extracted a larger amount of ancestry information in African Americans than previously reported SNP panels and provides nearly uniform coverage of the genome. Moreover, in the current study, simulations show that this more informative panel improves power for admixture mapping in African Americans when ethnicity risk ratios are modest. This is particularly important in the application of admixture mapping in complex genetic diseases for which only modest ethnicity risk ratios of relevant susceptibility genes are expected.
Figures
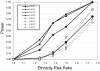
Figure 1.
δ of EURA/WAFR SNP AIMs. The SNP AIMs were arranged in ascending allele frequency in the AFA population, and, for each SNP, the WAFR SNP allele was chosen as the higher-frequency allele. Since AIM SNPs were chosen to maximize FIC, the majority of SNPs are close to fixation in the WAFR subjects. The data are based on nearly complete genotyping results in 84 EURA, 142 WAFR, and 96 AFA subjects.
Figure 2.
Admixture-mapping distribution for each chromosome. The admixture-mapping information (ordinate) is shown for each position on the deCODE sex-averaged map. The information was determined using ADMIXMAP analysis of genotyping results of 4,222 SNPs typed in the AFA samples (96 subjects).
Figure 3.
Power for admixture mapping as a function of admixture-mapping information. The power was determined from simulations with 700 cases, 700 controls, and SNP sets with admixture information corresponding to the legend for the SNP set used (see the “Analysis of Power Using Simulated Data Sets” and “Methods” sections). The power curves were determined using the ADMIXMAP program and deCODE genetic map for either case-only (CO) or case-control (CC) analyses. The appropriate α level for these analyses was a normalized score of 4.0, which was based on extensive simulations. The results are based on a minimum of 50 separate simulations and analysis for each measurement. Similar results were obtained using ANCESTRYMAP and MALDSOFT algorithms in more-limited analyses (data not shown).
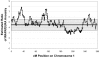
Figure 4.
Estimated ratio of WAFR/EURA ancestry across chromosome 1 in 96 AFA individuals. The ancestry ratio was determined by scoring each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For chromosome 1, a mean of 179 of a possible 192 chromosomes for each marker were scored as “WAFR” or “EURA” ancestry, with use of a log likelihood probability ratio >2. The figure shows the autosomal mean ±1 SD (gray rectangle).
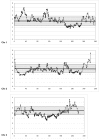
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Figure 5.
Estimated ratio of WAFR/EURA ancestry across each chromosome in 96 AFA individuals. The ratio was determined by scoring the ancestry of each marker on the basis of the assignment of ancestry (WAFR vs. EURA) in each predicted gamete (see the “Methods” section). For the autosomal chromosomes, a mean of 178 of a possible 192 chromosomes for each marker was scored as WAFR or EURA ancestry with use of a log likelihood probability ratio >2. For the autosomal chromosomes, the gray rectangle on each chromosome shows the autosomal mean ±1 SD (4.5±1.0). Similarly, for the X chromosome, the gray rectangle shows the mean ±1 SD (6.77±1.6). Many regions at the ends of chromosomes (qter and pter) show large deviations that correspond to decreased admixture information in these regions. The other deviations do not correlate with admixture information. As noted in the text, similar variations, although at different genomic positions, were found in simulations of 96 AFA individuals.
Similar articles
- A genomewide admixture mapping panel for Hispanic/Latino populations.
Mao X, Bigham AW, Mei R, Gutierrez G, Weiss KM, Brutsaert TD, Leon-Velarde F, Moore LG, Vargas E, McKeigue PM, Shriver MD, Parra EJ. Mao X, et al. Am J Hum Genet. 2007 Jun;80(6):1171-8. doi: 10.1086/518564. Epub 2007 Apr 20. Am J Hum Genet. 2007. PMID: 17503334 Free PMC article. - A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping.
Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, Silva G, Belmont JW, Hanson RL, Knowler WC, Gregersen PK, Ballinger DG, Seldin MF. Tian C, et al. Am J Hum Genet. 2007 Jun;80(6):1014-23. doi: 10.1086/513522. Am J Hum Genet. 2007. PMID: 17557415 Free PMC article. - Development of admixture mapping panels for African Americans from commercial high-density SNP arrays.
Chen G, Shriner D, Zhou J, Doumatey A, Huang H, Gerry NP, Herbert A, Christman MF, Chen Y, Dunston GM, Faruque MU, Rotimi CN, Adeyemo A. Chen G, et al. BMC Genomics. 2010 Jul 5;11:417. doi: 10.1186/1471-2164-11-417. BMC Genomics. 2010. PMID: 20602785 Free PMC article. - African Americans with asthma: genetic insights.
Barnes KC, Grant AV, Hansel NN, Gao P, Dunston GM. Barnes KC, et al. Proc Am Thorac Soc. 2007 Jan;4(1):58-68. doi: 10.1513/pats.200607-146JG. Proc Am Thorac Soc. 2007. PMID: 17202293 Free PMC article. Review. - Genetic admixture: a tool to identify diabetic nephropathy genes in African Americans.
Divers J, Moossavi S, Langefeld CD, Freedman BI. Divers J, et al. Ethn Dis. 2008 Summer;18(3):384-8. Ethn Dis. 2008. PMID: 18785456 Review.
Cited by
- Ancestry inference in complex admixtures via variable-length Markov chain linkage models.
Rodriguez JM, Bercovici S, Elmore M, Batzoglou S. Rodriguez JM, et al. J Comput Biol. 2013 Mar;20(3):199-211. doi: 10.1089/cmb.2012.0088. Epub 2013 Feb 19. J Comput Biol. 2013. PMID: 23421795 Free PMC article. - A high-density admixture scan in 1,670 African Americans with hypertension.
Deo RC, Patterson N, Tandon A, McDonald GJ, Haiman CA, Ardlie K, Henderson BE, Henderson SO, Reich D. Deo RC, et al. PLoS Genet. 2007 Nov;3(11):e196. doi: 10.1371/journal.pgen.0030196. PLoS Genet. 2007. PMID: 18020707 Free PMC article. - Type 2 deiodinase and host responses of sepsis and acute lung injury.
Ma SF, Xie L, Pino-Yanes M, Sammani S, Wade MS, Letsiou E, Siegler J, Wang T, Infusino G, Kittles RA, Flores C, Zhou T, Prabhakar BS, Moreno-Vinasco L, Villar J, Jacobson JR, Dudek SM, Garcia JG. Ma SF, et al. Am J Respir Cell Mol Biol. 2011 Dec;45(6):1203-11. doi: 10.1165/rcmb.2011-0179OC. Epub 2011 Jun 17. Am J Respir Cell Mol Biol. 2011. PMID: 21685153 Free PMC article. - Applying genome-wide gene-based expression quantitative trait locus mapping to study population ancestry and pharmacogenetics.
Yang HC, Lin CW, Chen CW, Chen JJ. Yang HC, et al. BMC Genomics. 2014 Apr 29;15:319. doi: 10.1186/1471-2164-15-319. BMC Genomics. 2014. PMID: 24779372 Free PMC article. - Race, genetic West African ancestry, and prostate cancer prediction by prostate-specific antigen in prospectively screened high-risk men.
Giri VN, Egleston B, Ruth K, Uzzo RG, Chen DY, Buyyounouski M, Raysor S, Hooker S, Torres JB, Ramike T, Mastalski K, Kim TY, Kittles R. Giri VN, et al. Cancer Prev Res (Phila). 2009 Mar;2(3):244-50. doi: 10.1158/1940-6207.CAPR-08-0150. Epub 2009 Feb 24. Cancer Prev Res (Phila). 2009. PMID: 19240249 Free PMC article.
References
Web Resources
- Institut für Humangenetik, http://ihg.gsf.de/cgi-bin/hw/hwa1.pl (for the FINETTI software)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for lupus, prostate cancer, diabetic nephropathy, multiple sclerosis, and osteoporosis)
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous