Ultra-high resolution crystal structure of HIV-1 protease mutant reveals two binding sites for clinical inhibitor TMC114 - PubMed (original) (raw)
Ultra-high resolution crystal structure of HIV-1 protease mutant reveals two binding sites for clinical inhibitor TMC114
Andrey Y Kovalevsky et al. J Mol Biol. 2006.
Erratum in
- J Mol Biol. 2007 Jan 19;365(3):901
Abstract
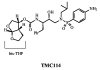
TMC114 (darunavir) is a promising clinical inhibitor of HIV-1 protease (PR) for treatment of drug resistant HIV/AIDS. We report the ultra-high 0.84 A resolution crystal structure of the TMC114 complex with PR containing the drug-resistant mutation V32I (PR(V32I)), and the 1.22 A resolution structure of a complex with PR(M46L). These structures show TMC114 bound at two distinct sites, one in the active-site cavity and the second on the surface of one of the flexible flaps in the PR dimer. Remarkably, TMC114 binds at these two sites simultaneously in two diastereomers related by inversion of the sulfonamide nitrogen. Moreover, the flap site is shaped to accommodate the diastereomer with the S-enantiomeric nitrogen rather than the one with the R-enantiomeric nitrogen. The existence of the second binding site and two diastereomers suggest a mechanism for the high effectiveness of TMC114 on drug-resistant HIV and the potential design of new inhibitors.
Figures
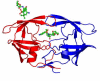
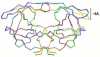
Figure 1
(a) PRV32I dimer structure. Two subunits (in red and blue) are shown indicating the secondary structure. TMC114 is in ball-and-stick representation colored by atom type, and is bound in two sites. (b) and (c) The electron density (2FO-FC) for residues 55-60 in the PRV32I and PRM46L structures. Contour levels are 3.6σ for (b) and 2.4σ for (c). (d) The 2FO-FC electron density for TMC114 bound to the flap in PRV32I contoured at 1.8σ. TMC114 has 60% occupancy, while the other 40% correspond to a DMSO solvent molecule, depicted in magenta. (e) Structures of TMC114 bound in the active site cavity (_R_-enantiomer) and in the flap region (_S_-enantiomer). The moieties in the box are related by reflection in a mirror and can be obtained by inversion of the sulfonamide nitrogen.
Figure 1
(a) PRV32I dimer structure. Two subunits (in red and blue) are shown indicating the secondary structure. TMC114 is in ball-and-stick representation colored by atom type, and is bound in two sites. (b) and (c) The electron density (2FO-FC) for residues 55-60 in the PRV32I and PRM46L structures. Contour levels are 3.6σ for (b) and 2.4σ for (c). (d) The 2FO-FC electron density for TMC114 bound to the flap in PRV32I contoured at 1.8σ. TMC114 has 60% occupancy, while the other 40% correspond to a DMSO solvent molecule, depicted in magenta. (e) Structures of TMC114 bound in the active site cavity (_R_-enantiomer) and in the flap region (_S_-enantiomer). The moieties in the box are related by reflection in a mirror and can be obtained by inversion of the sulfonamide nitrogen.
Figure 1
(a) PRV32I dimer structure. Two subunits (in red and blue) are shown indicating the secondary structure. TMC114 is in ball-and-stick representation colored by atom type, and is bound in two sites. (b) and (c) The electron density (2FO-FC) for residues 55-60 in the PRV32I and PRM46L structures. Contour levels are 3.6σ for (b) and 2.4σ for (c). (d) The 2FO-FC electron density for TMC114 bound to the flap in PRV32I contoured at 1.8σ. TMC114 has 60% occupancy, while the other 40% correspond to a DMSO solvent molecule, depicted in magenta. (e) Structures of TMC114 bound in the active site cavity (_R_-enantiomer) and in the flap region (_S_-enantiomer). The moieties in the box are related by reflection in a mirror and can be obtained by inversion of the sulfonamide nitrogen.
Figure 1
(a) PRV32I dimer structure. Two subunits (in red and blue) are shown indicating the secondary structure. TMC114 is in ball-and-stick representation colored by atom type, and is bound in two sites. (b) and (c) The electron density (2FO-FC) for residues 55-60 in the PRV32I and PRM46L structures. Contour levels are 3.6σ for (b) and 2.4σ for (c). (d) The 2FO-FC electron density for TMC114 bound to the flap in PRV32I contoured at 1.8σ. TMC114 has 60% occupancy, while the other 40% correspond to a DMSO solvent molecule, depicted in magenta. (e) Structures of TMC114 bound in the active site cavity (_R_-enantiomer) and in the flap region (_S_-enantiomer). The moieties in the box are related by reflection in a mirror and can be obtained by inversion of the sulfonamide nitrogen.
Figure 1
(a) PRV32I dimer structure. Two subunits (in red and blue) are shown indicating the secondary structure. TMC114 is in ball-and-stick representation colored by atom type, and is bound in two sites. (b) and (c) The electron density (2FO-FC) for residues 55-60 in the PRV32I and PRM46L structures. Contour levels are 3.6σ for (b) and 2.4σ for (c). (d) The 2FO-FC electron density for TMC114 bound to the flap in PRV32I contoured at 1.8σ. TMC114 has 60% occupancy, while the other 40% correspond to a DMSO solvent molecule, depicted in magenta. (e) Structures of TMC114 bound in the active site cavity (_R_-enantiomer) and in the flap region (_S_-enantiomer). The moieties in the box are related by reflection in a mirror and can be obtained by inversion of the sulfonamide nitrogen.
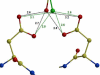
Figure 2
Hydrogen bonds between the central OH group of TMC114 and the catalytic Asp25 and Asp25'. The major conformation of TMC114 is colored by atom type, and the minor conformation is green. Interatomic distances are shown in Å (a) PR-TMC114 (PDB code 1S6G); the TMC114 conformations were refined with 55% and 45% occupancies. (b) PRV32I–TMC114 and (c) PRM46L–TMC114. The 2FO-FC electron density for the active site residues Asp25 and Asp25' is shown with the contour levels of 2.2σ. The alternate conformations have occupancies of 60% and 40%.
Figure 2
Hydrogen bonds between the central OH group of TMC114 and the catalytic Asp25 and Asp25'. The major conformation of TMC114 is colored by atom type, and the minor conformation is green. Interatomic distances are shown in Å (a) PR-TMC114 (PDB code 1S6G); the TMC114 conformations were refined with 55% and 45% occupancies. (b) PRV32I–TMC114 and (c) PRM46L–TMC114. The 2FO-FC electron density for the active site residues Asp25 and Asp25' is shown with the contour levels of 2.2σ. The alternate conformations have occupancies of 60% and 40%.
Figure 2
Hydrogen bonds between the central OH group of TMC114 and the catalytic Asp25 and Asp25'. The major conformation of TMC114 is colored by atom type, and the minor conformation is green. Interatomic distances are shown in Å (a) PR-TMC114 (PDB code 1S6G); the TMC114 conformations were refined with 55% and 45% occupancies. (b) PRV32I–TMC114 and (c) PRM46L–TMC114. The 2FO-FC electron density for the active site residues Asp25 and Asp25' is shown with the contour levels of 2.2σ. The alternate conformations have occupancies of 60% and 40%.
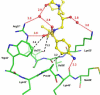
Figure 3
Hydrogen bond, C-H…O and C-H…π interactions are shown in the active site cavity of PRV32I for the major conformation of TMC114 (a) and the minor conformation (b). Interactions for the alternate conformations of TMC114 in PR and PRM46L are shown in the Supplementary Material.
Figure 3
Hydrogen bond, C-H…O and C-H…π interactions are shown in the active site cavity of PRV32I for the major conformation of TMC114 (a) and the minor conformation (b). Interactions for the alternate conformations of TMC114 in PR and PRM46L are shown in the Supplementary Material.
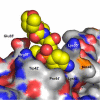
Figure 4
(a) TMC114 at the flap binding site in PRV32I, and (b) similar view in PR. TMC114 in the PRV32I complex, and a glycerol molecule in PR-TMC114 are in a space-filling representation and colored by atom type. The protease is represented as a surface, and the residues forming the binding site are labeled. (c) Superposition of _R_-enantiomer (magenta) from the active-site cavity with the _S_-enantiomer (colored by atom type) bound in the flap site of PRV32I. The aniline moiety of the R-enantiomer (indicated by arrow) collides with the protease residues, which would prevent it from binding in the flap site. The geometry is similar in PRM46L.
Figure 4
(a) TMC114 at the flap binding site in PRV32I, and (b) similar view in PR. TMC114 in the PRV32I complex, and a glycerol molecule in PR-TMC114 are in a space-filling representation and colored by atom type. The protease is represented as a surface, and the residues forming the binding site are labeled. (c) Superposition of _R_-enantiomer (magenta) from the active-site cavity with the _S_-enantiomer (colored by atom type) bound in the flap site of PRV32I. The aniline moiety of the R-enantiomer (indicated by arrow) collides with the protease residues, which would prevent it from binding in the flap site. The geometry is similar in PRM46L.
Figure 4
(a) TMC114 at the flap binding site in PRV32I, and (b) similar view in PR. TMC114 in the PRV32I complex, and a glycerol molecule in PR-TMC114 are in a space-filling representation and colored by atom type. The protease is represented as a surface, and the residues forming the binding site are labeled. (c) Superposition of _R_-enantiomer (magenta) from the active-site cavity with the _S_-enantiomer (colored by atom type) bound in the flap site of PRV32I. The aniline moiety of the R-enantiomer (indicated by arrow) collides with the protease residues, which would prevent it from binding in the flap site. The geometry is similar in PRM46L.
Figure 5
. (a) Hydrogen bond network and C-H…O interactions of TMC114 bound in the flap site of PRV32I. Hydrogen bonds are colored in red, C-H…O contacts are black, and distances are in Å The interactions in the PRM46L complex are very similar. (b) TMC114 bound to the surface site is surrounded by four protein molecules. The asymmetric unit consists of PRV32I (blue) and two inhibitor molecules shown in yellow ball-and-stick representations. The symmetry related protease molecules are in cyan, orange and magenta.
Figure 5
. (a) Hydrogen bond network and C-H…O interactions of TMC114 bound in the flap site of PRV32I. Hydrogen bonds are colored in red, C-H…O contacts are black, and distances are in Å The interactions in the PRM46L complex are very similar. (b) TMC114 bound to the surface site is surrounded by four protein molecules. The asymmetric unit consists of PRV32I (blue) and two inhibitor molecules shown in yellow ball-and-stick representations. The symmetry related protease molecules are in cyan, orange and magenta.
Figure 6
(a) The superposition of the PRV32I and PRM46L structures onto the wild type PR. (b) The residues (labeled) of the flap binding site for TMC114 have the largest differences. PR is colored by atom type, while PRV32I and PRM46L are colored in magenta and cyan, respectively. The atomic shifts (Å) are indicated by dashed arrows.
Figure 6
(a) The superposition of the PRV32I and PRM46L structures onto the wild type PR. (b) The residues (labeled) of the flap binding site for TMC114 have the largest differences. PR is colored by atom type, while PRV32I and PRM46L are colored in magenta and cyan, respectively. The atomic shifts (Å) are indicated by dashed arrows.
Similar articles
- Exploring the drug resistance of V32I and M46L mutant HIV-1 protease to inhibitor TMC114: flap dynamics and binding mechanism.
Meher BR, Wang Y. Meher BR, et al. J Mol Graph Model. 2015 Mar;56:60-73. doi: 10.1016/j.jmgm.2014.11.003. Epub 2014 Dec 5. J Mol Graph Model. 2015. PMID: 25562662 Free PMC article. - Interaction of I50V mutant and I50L/A71V double mutant HIV-protease with inhibitor TMC114 (darunavir): molecular dynamics simulation and binding free energy studies.
Meher BR, Wang Y. Meher BR, et al. J Phys Chem B. 2012 Feb 16;116(6):1884-900. doi: 10.1021/jp2074804. Epub 2012 Feb 3. J Phys Chem B. 2012. PMID: 22239286 Free PMC article. - High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains.
Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. Tie Y, et al. J Mol Biol. 2004 Apr 23;338(2):341-52. doi: 10.1016/j.jmb.2004.02.052. J Mol Biol. 2004. PMID: 15066436 - Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.
Lefebvre E, Schiffer CA. Lefebvre E, et al. AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review. - Darunavir, a conceptually new HIV-1 protease inhibitor for the treatment of drug-resistant HIV.
Ghosh AK, Dawson ZL, Mitsuya H. Ghosh AK, et al. Bioorg Med Chem. 2007 Dec 15;15(24):7576-80. doi: 10.1016/j.bmc.2007.09.010. Epub 2007 Sep 14. Bioorg Med Chem. 2007. PMID: 17900913 Free PMC article. Review.
Cited by
- The good, the bad and the twisted: a survey of ligand geometry in protein crystal structures.
Liebeschuetz J, Hennemann J, Olsson T, Groom CR. Liebeschuetz J, et al. J Comput Aided Mol Des. 2012 Feb;26(2):169-83. doi: 10.1007/s10822-011-9538-6. Epub 2012 Jan 14. J Comput Aided Mol Des. 2012. PMID: 22246295 Free PMC article. - Structural Insights to Human Immunodeficiency Virus (HIV-1) Targets and Their Inhibition.
Vanangamudi M, Nair PC, Engels SEM, Palaniappan S, Namasivayam V. Vanangamudi M, et al. Adv Exp Med Biol. 2021;1322:63-95. doi: 10.1007/978-981-16-0267-2_3. Adv Exp Med Biol. 2021. PMID: 34258737 - Capturing the reaction pathway in near-atomic-resolution crystal structures of HIV-1 protease.
Shen CH, Tie Y, Yu X, Wang YF, Kovalevsky AY, Harrison RW, Weber IT. Shen CH, et al. Biochemistry. 2012 Oct 2;51(39):7726-32. doi: 10.1021/bi3008092. Epub 2012 Sep 21. Biochemistry. 2012. PMID: 22963370 Free PMC article. - Current and Novel Inhibitors of HIV Protease.
Pokorná J, Machala L, Rezáčová P, Konvalinka J. Pokorná J, et al. Viruses. 2009 Dec;1(3):1209-39. doi: 10.3390/v1031209. Epub 2009 Dec 11. Viruses. 2009. PMID: 21994591 Free PMC article. - Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease.
Kovalevsky AY, Louis JM, Aniana A, Ghosh AK, Weber IT. Kovalevsky AY, et al. J Mol Biol. 2008 Dec 5;384(1):178-92. doi: 10.1016/j.jmb.2008.09.031. Epub 2008 Sep 20. J Mol Biol. 2008. PMID: 18834890 Free PMC article.
References
- De Clercq E. New Approaches toward Anti-HIV Chemotherapy. J. Med. Chem. 2005;48:1–17. - PubMed
- Barlett JA, DeMasi R, Quinn J, Moxham C, Rousseau F. Overview of the Effectiveness of Triple Combination Therapy in Antiretroviral-Naïve HIV-1 Infected Adults. AIDS. 2001;15:1369–1377. - PubMed
- Gulick RM, Mellors JW, Havlir D, Eron JJ, Meibohm A, Condra JH, Valentine FT, McMahon D, Gonzalez C, Jonas L, Emini EA, Chodakewitz JA, Isaacs R, Richman DD. 3-Year Suppression if HIV Viremia with Indinavir, Zidovudine, and Lamivudine. Ann. Intern. Med. 2000;133:35–39. - PubMed
- Wlodawer A, Vondrasek J. Inhibitors of HIV-1 Protease: A Major Success of Structure-Assisted Drug Design. Ann. Rev. Biophys. Biomol. Struct. 1998;27:249–284. - PubMed
- Hertogs K, Bloor S, Kemp SD, Van den Eynde C, Alcorn TM, Pauwels R, Van Houtte M, Staszewski S, Miller V, Larder BA. Phenotypic and Genotypic Analysis of Clinical HIV-1 Isolates Reveals Extensive Protease Inhibitor Cross-resistance: A Survey of Over 6000 Samples. AIDS. 2000;14:1203–1210. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM62920/GM/NIGMS NIH HHS/United States
- U01 GM062920/GM/NIGMS NIH HHS/United States
- R01 GM062920/GM/NIGMS NIH HHS/United States
- R03 TW001001/TW/FIC NIH HHS/United States
- Intramural NIH HHS/United States
- GM53386/GM/NIGMS NIH HHS/United States
- R37 GM053386/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials