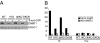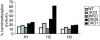Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival - PubMed (original) (raw)
Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival
Gerda Egger et al. Proc Natl Acad Sci U S A. 2006.
Abstract
Previous studies have shown that DNA methyltransferase (Dnmt) 1 is required for maintenance of bulk DNA methylation and is essential for mouse development. However, somatic disruption of DNMT1 in the human cancer cell line HCT116 was not lethal and caused only minor decreases in methylation. Here, we report the identification of a truncated DNMT1 protein, which was generated by the disruption of DNMT1 in HCT116 cells. The truncated protein, which had parts of the regulatory N-terminal domain deleted but preserved the catalytic C-terminal domain, was present at different levels in all DNMT1 single-knockout and DNMT1/DNMT3b double-knockout cell lines tested and retained hemimethylase activity. DNMT1 RNAi resulted in decreased cell viability in WT and knockout cells and further loss of DNA methylation in DNMT1 knockout cells. Furthermore, we observed a delay in methylation after replication and an increase in hemimethylation of specific CpG sites in cells expressing the truncated protein. Remethylation studies after drug-induced hypomethylation suggest a putative role of DNMT1 in the de novo methylation of a subtelomeric repeat, D4Z4, which is lost in cells lacking full-length DNMT1. Our data suggest that DNMT1 might be essential for maintenance of DNA methylation, proliferation, and survival of cancer cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
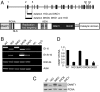
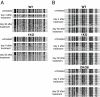
DNMT1 expression in different HCT116 cell lines. (A Upper) Genomic map of WT DNMT1. Black vertical boxes indicate exons; black horizontal bars drawn below indicate deletions determined by direct sequencing of transcripts in different knockout cell lines. (A Lower) DNMT1 protein domain structure. DMAP1 domain, amino acids 1–120; PCNA domain, amino acids 163–174; NLS, nuclear localization signal domain, amino acids 177–205; DNA replication foci-targeting domain, amino acids 331–550; ZN D, zinc finger region, amino acids 646–692; KEN, KEN box (KENxxxR), amino acids 644–650; BAH1 and BAH2, bromo-adjacent homology domains, amino acids 755–880 and 972–1100, respectively; KG, 6 × 2-aa tandem repeats of K-G, amino acids 1109–1120; catalytic domain, amino acids 1139–1616. Note that illustrations are not drawn to scale. (B) RT-PCR in different WT and knockout cell lines was performed by using primers located in exons 1 and 6 of WT DNMT1 (E1–6), half exons 1 and 6 and exon 10 of DNMT1 (E1/6–10), or exons 32 and 34 (E32–34). As a control, primers against β-actin were used (Actin). (C) Western blot analysis of various HCT116 cell lines with C-terminal DNMT1 (Upper) or PCNA antibody as a loading control (Lower). (D) Quantitation of the Western blot shown in C for DNMT1 protein levels, normalized to PCNA expression. The WT expression level was arbitrarily set as 1. WT, HCT116 WT; 1KO, _DNMT1_−/− cells; 3bKO, _DNMT3b_−/− cells; DKO8 and DKO1, two independent DNMT1 and DNMT3b DKO clones, respectively.
Fig. 2.
DNMT1 activity. (A) Western blot analysis of cells treated with 0.3 μM 5-aza-CdR or left untreated for 24 h. The blot was sequentially probed with an antibody against the C terminus of DNMT1 and HDAC1 as a loading control. (B) DNMT1 was immunoprecipitated with the same DNMT1 antibody as in A and incubated with hemimethylated or unmethylated DNA fragments corresponding to a 428-bp-long p16 intron 1 sequence and 160 μM _S_-adenosylmethionine for 3 h at 37°C. The percentage of methylation was determined by quantitative Ms-SNuPE (33). IP, immunoprecipitation with DNMT1 antibody; Pre, control immunoprecipitation with whole rabbit serum.
Fig. 3.
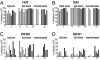
DNMT1 siRNA. (A) Western blot of HCT116 WT and knockout cells untransfected (0) or transiently transfected with negative control siRNA (C) or DNMT1 siRNA (RNAi) and probed with a DNMT1 or PCNA antibody. (B) Percentage of viability was calculated as the ratio of the number of DNMT1 RNAi-transfected cells to control-transfected cells. (C) Ms-SNuPE assays of untransfected, control, and DNMT1 RNAi cells for different loci. D4Z4, subtelomeric repeat, CpG island; RUNX1, CpG-poor region in intron 1; MAGE A1, CpG-poor promoter region; TIMP3, CpG island promoter region. Data shown are representatives of at least three independently repeated experiments. NS, P value not significant; *, P value significant; T, P value shows a tendency toward significance.
Fig. 4.
Hemimethylation assay. Hemimethylation was assessed in the different HCT116 cell lines at three different sites within the p16 gene (H1, H2, and H3). H1 is located in the promoter, H2 is in a CpG-poor region within intron 1, and H3 is in a CpG-rich region of intron 1. Percentage of hemimethylation was defined as h/(h + f), where h represents hemimethylated sites (only one strand methylated) and f represents fully methylated sites (both strands methylated).
Fig. 5.
Methylation of various regions in HCT116 WT and knockout cells. Methylation levels (average of two to three CpG sites at each locus) were determined by Ms-SNuPE analyses and graphed as a percentage of WT methylation levels. Open bars represent promoter regions, and filled bars indicate nonpromoter regions. The numbering of regions is as follows. CpG-poor regions: 1, RUNX3 promoter; 2, MAGE A1 promoter; 3, NANOG promoter; 4, p16 intron 1; 5, PAX3 intron; 6, RUNX1 intron; 7, BDNF intron; 8, PCNA intron. CpG islands: 9, p16 promoter; 10, ENDRB promoter; 11, ATBF1 promoter; 12, XIST promoter; 13, TIMP3 promoter; 14, TPEF promoter; 15, DNMT3a2 promoter; 16, microRNA-127; 17, p16 exon 1; 18, p16 intron 4; 19, _M4_-4 single-copy sequence, chromosome 16q22; 20, p16 exon 2. Imprinted regions: 21, H19; 22, SNRPN. Repeats: 23, D4Z4; 24, p53-Alu; 25, p16-Alu; 26, LINE global; 27, Alu global.
Fig. 6.
Methylation kinetics of newly synthesized DNA. The different cell lines were pulsed for 1 h with BrdU and subsequently chased with thymidine for 0, 15, or 30 min or 8 h. DNA that had incorporated BrdU was isolated by anti-BrdU immunoprecipitation. Methylation levels were determined by Ms-SNuPE analyses at the indicated time points. (A) Methylation kinetics of a CpG-poor region within the RUNX1 intron. (B) Methylation levels of the CpG-rich subtelomeric repeat D4Z4 at different time points of the pulse–chase.
Fig. 7.
Remethylation after 5-aza-CdR treatment. HCT116 WT and knockout cells were treated with 0.3 μM 5-aza-CdR for 24 h and released from the drug thereafter. Methylation levels were determined by bisulfite sequencing in untreated cells and on days 4 and 32 after treatment. Each line with circles indicates an individual DNA molecule. Open circles represent unmethylated cytosines, and filled circles represent methylated cytosines within a CpG dinucleotide context. (A) Bisulfite sequencing of D4Z4. (B) Sequencing results for a p16 exon 2 region.
Similar articles
- Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b.
James SR, Link PA, Karpf AR. James SR, et al. Oncogene. 2006 Nov 2;25(52):6975-85. doi: 10.1038/sj.onc.1209678. Epub 2006 May 22. Oncogene. 2006. PMID: 16715135 - Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation.
Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Weisenberger DJ, et al. Mol Cancer Res. 2004 Jan;2(1):62-72. Mol Cancer Res. 2004. PMID: 14757847 - DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells.
Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Längst G, Leonhardt H. Spada F, et al. J Cell Biol. 2007 Feb 26;176(5):565-71. doi: 10.1083/jcb.200610062. Epub 2007 Feb 20. J Cell Biol. 2007. PMID: 17312023 Free PMC article. - DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells.
Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. Robert MF, et al. Nat Genet. 2003 Jan;33(1):61-5. doi: 10.1038/ng1068. Epub 2002 Dec 23. Nat Genet. 2003. PMID: 12496760 - Non-catalytic functions of DNMT1.
Espada J. Espada J. Epigenetics. 2012 Feb;7(2):115-8. doi: 10.4161/epi.7.2.18756. Epigenetics. 2012. PMID: 22395459 Review.
Cited by
- DNA methylation governs the sensitivity of repeats to restriction by the HUSH-MORC2 corepressor.
Pandiloski N, Horváth V, Karlsson O, Koutounidou S, Dorazehi F, Christoforidou G, Matas-Fuentes J, Gerdes P, Garza R, Jönsson ME, Adami A, Atacho DAM, Johansson JG, Englund E, Kokaia Z, Jakobsson J, Douse CH. Pandiloski N, et al. Nat Commun. 2024 Aug 30;15(1):7534. doi: 10.1038/s41467-024-50765-4. Nat Commun. 2024. PMID: 39214989 Free PMC article. - Select EZH2 inhibitors enhance viral mimicry effects of DNMT inhibition through a mechanism involving NFAT:AP-1 signaling.
Chomiak AA, Tiedemann RL, Liu Y, Kong X, Cui Y, Wiseman AK, Thurlow KE, Cornett EM, Topper MJ, Baylin SB, Rothbart SB. Chomiak AA, et al. Sci Adv. 2024 Mar 29;10(13):eadk4423. doi: 10.1126/sciadv.adk4423. Epub 2024 Mar 27. Sci Adv. 2024. PMID: 38536911 Free PMC article. - Comprehensive Evaluation of The Infinium Human MethylationEPIC v2 BeadChip.
Kaur D, Lee SM, Goldberg D, Spix NJ, Hinoue T, Li HT, Dwaraka VB, Smith R, Shen H, Liang G, Renke N, Laird PW, Zhou W. Kaur D, et al. Epigenetics Commun. 2023;3(1):6. doi: 10.1186/s43682-023-00021-5. Epub 2023 Sep 27. Epigenetics Commun. 2023. PMID: 38455390 Free PMC article. - Tunable DNMT1 degradation reveals DNMT1/DNMT3B synergy in DNA methylation and genome organization.
Scelfo A, Barra V, Abdennur N, Spracklin G, Busato F, Salinas-Luypaert C, Bonaiti E, Velasco G, Bonhomme F, Chipont A, Tijhuis AE, Spierings DCJ, Guérin C, Arimondo P, Francastel C, Foijer F, Tost J, Mirny L, Fachinetti D. Scelfo A, et al. J Cell Biol. 2024 Apr 1;223(4):e202307026. doi: 10.1083/jcb.202307026. Epub 2024 Feb 20. J Cell Biol. 2024. PMID: 38376465 Free PMC article. - Flow cytometry of DNMT1 as a biomarker of hypomethylating therapies.
Woost PG, William BM, Cooper BW, Ueda Oshima M, Otegbeye F, De Lima MJ, Wald D, Mahfouz RZ, Saunthararajah Y, Stefan T, Jacobberger JW. Woost PG, et al. Cytometry B Clin Cytom. 2024 Jan;106(1):11-24. doi: 10.1002/cyto.b.22158. Epub 2024 Feb 12. Cytometry B Clin Cytom. 2024. PMID: 38345160
References
- Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481–514. - PubMed
- Li E, Bestor TH, Jaenisch R. Cell. 1992;69:915–926. - PubMed
- Li E, Beard C, Forster AC, Bestor TH, Jaenisch R. Cold Spring Harbor Symp Quant Biol. 1993;58:297–305. - PubMed
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Nat Genet. 2001;27:31–39. - PubMed
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Cell. 1995;81:197–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5251143701/PHS HHS/United States
- R01 CA 82422/CA/NCI NIH HHS/United States
- R01 CA082422/CA/NCI NIH HHS/United States
- R01 CA 83867/CA/NCI NIH HHS/United States
- R01 CA083867/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources