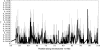Estimating recombination rates from single-nucleotide polymorphisms using summary statistics - PubMed (original) (raw)
Comparative Study
Estimating recombination rates from single-nucleotide polymorphisms using summary statistics
Badri Padhukasahasram et al. Genetics. 2006 Nov.
Abstract
We describe a novel method for jointly estimating crossing-over and gene-conversion rates from population genetic data using summary statistics. The performance of our method was tested on simulated data sets and compared with the composite-likelihood method of R. R. Hudson. For several realistic parameter values, the new method performed similarly to the composite-likelihood approach for estimating crossing-over rates and better when estimating gene-conversion rates. We used our method to analyze a human data set recently genotyped by Perlegen Sciences.
Figures
Figure 1.—
Recombination rate estimates for 50-kb overlapping windows across a 50-Mb region in chromosome 1. (Top) The population gene-conversion rates and (bottom) the population crossing-over rates, estimated under a model with uniform recombination for a mean conversion tract length (L) of 500 bp. The shaded bars represent confidence intervals with at least 90% of the total mass and are constructed around the estimated values at positions corresponding to the centers of the windows.
Figure 1.—
Recombination rate estimates for 50-kb overlapping windows across a 50-Mb region in chromosome 1. (Top) The population gene-conversion rates and (bottom) the population crossing-over rates, estimated under a model with uniform recombination for a mean conversion tract length (L) of 500 bp. The shaded bars represent confidence intervals with at least 90% of the total mass and are constructed around the estimated values at positions corresponding to the centers of the windows.
Figure 2.—
Comparison of recombination rates estimated from population genetic data using the summary statistics method with those obtained from human genetic maps (data from K
ong
et al. 2002 used in M
yers
et al. 2005). A scatter plot is shown of the estimates of population-scaled crossing-over rate with the deCODE pedigree-based estimates (centimorgans per megabase) for 5-Mb regions across the human genome. The correlation coefficient between these estimates is 0.9181.
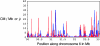
Figure 3.—
Crossing-over estimates in a 3.3-Mb MHC region in human chromosome 6. The red curve shows estimates (centimorgans per megabase) obtained in M
yers
et al. (2005) using the M
c
V
ean
et al. (2004) method. The blue curve shows sliding-window population crossing-over estimates (multiplied by some constant) for overlapping 50-kb regions along chromosome 6 obtained using the summary statistics method.
Similar articles
- Estimating recombination rates using three-site likelihoods.
Wall JD. Wall JD. Genetics. 2004 Jul;167(3):1461-73. doi: 10.1534/genetics.103.025742. Genetics. 2004. PMID: 15280255 Free PMC article. - Estimating the rate of gene conversion on human chromosome 21.
Padhukasahasram B, Marjoram P, Nordborg M. Padhukasahasram B, et al. Am J Hum Genet. 2004 Sep;75(3):386-97. doi: 10.1086/423451. Epub 2004 Jul 12. Am J Hum Genet. 2004. PMID: 15250027 Free PMC article. - A comparison of three estimators of the population-scaled recombination rate: accuracy and robustness.
Smith NG, Fearnhead P. Smith NG, et al. Genetics. 2005 Dec;171(4):2051-62. doi: 10.1534/genetics.104.036293. Epub 2005 Jun 14. Genetics. 2005. PMID: 15956675 Free PMC article. - Justified chauvinism: advances in defining meiotic recombination through sperm typing.
Carrington M, Cullen M. Carrington M, et al. Trends Genet. 2004 Apr;20(4):196-205. doi: 10.1016/j.tig.2004.02.006. Trends Genet. 2004. PMID: 15041174 Review. - Estimating recombination rates from population-genetic data.
Stumpf MP, McVean GA. Stumpf MP, et al. Nat Rev Genet. 2003 Dec;4(12):959-68. doi: 10.1038/nrg1227. Nat Rev Genet. 2003. PMID: 14631356 Review.
Cited by
- Impermanence of bacterial clones.
Bobay LM, Traverse CC, Ochman H. Bobay LM, et al. Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):8893-900. doi: 10.1073/pnas.1501724112. Proc Natl Acad Sci U S A. 2015. PMID: 26195749 Free PMC article. - Bayesian population genomic inference of crossing over and gene conversion.
Padhukasahasram B, Rannala B. Padhukasahasram B, et al. Genetics. 2011 Oct;189(2):607-19. doi: 10.1534/genetics.111.130195. Epub 2011 Aug 11. Genetics. 2011. PMID: 21840857 Free PMC article. - Joint estimation of gene conversion rates and mean conversion tract lengths from population SNP data.
Yin J, Jordan MI, Song YS. Yin J, et al. Bioinformatics. 2009 Jun 15;25(12):i231-9. doi: 10.1093/bioinformatics/btp229. Bioinformatics. 2009. PMID: 19477993 Free PMC article. - Recombination yet inefficient selection along the Drosophila melanogaster subgroup's fourth chromosome.
Arguello JR, Zhang Y, Kado T, Fan C, Zhao R, Innan H, Wang W, Long M. Arguello JR, et al. Mol Biol Evol. 2010 Apr;27(4):848-61. doi: 10.1093/molbev/msp291. Epub 2009 Dec 14. Mol Biol Evol. 2010. PMID: 20008457 Free PMC article. - Unraveling recombination rate evolution using ancestral recombination maps.
Munch K, Schierup MH, Mailund T. Munch K, et al. Bioessays. 2014 Sep;36(9):892-900. doi: 10.1002/bies.201400047. Epub 2014 Jul 14. Bioessays. 2014. PMID: 25043668 Free PMC article. Review.
References
- Allers, T., and M. Litchen, 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. - PubMed
- Crawford, D. C., T. Bhangale, N. Li, G. Hellenthal, M. J. Rieder et al., 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36: 700–706. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources