The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins - PubMed (original) (raw)
The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins
Mladen Tomich et al. J Bacteriol. 2006 Oct.
Abstract
The tad locus of Actinobacillus actinomycetemcomitans encodes genes for the biogenesis of Flp pili, which allow the bacterium to adhere tenaciously to surfaces and form strong biofilms. Although tad (tight adherence) loci are widespread among bacterial and archaeal species, very little is known about the functions of the individual components of the Tad secretion apparatus. Here we characterize the mechanism by which the pre-Flp1 prepilin is processed to the mature pilus subunit. We demonstrate that the tadV gene encodes a prepilin peptidase that is both necessary and sufficient for proteolytic maturation of Flp1. TadV was also found to be required for maturation of the TadE and TadF pilin-like proteins, which we term pseudopilins. Using site-directed mutagenesis, we show that processing of pre-Flp1, pre-TadE, and pre-TadF is required for biofilm formation. Mutation of a highly conserved glutamic acid residue at position +5 of Flp1, relative to the cleavage site, resulted in a processed pilin that was blocked in assembly. In contrast, identical mutations in TadE or TadF had no effect on biofilm formation, indicating that the mechanisms by which Flp1 pilin and the pseudopilins function are distinct. We also determined that two conserved aspartic acid residues in TadV are critical for function of the prepilin peptidase. Together, our results indicate that the A. actinomycetemcomitans TadV protein is a member of a novel subclass of nonmethylating aspartic acid prepilin peptidases.
Figures
FIG. 1.
The tad locus of A. actinomycetemcomitans. The position of each gene and the direction of transcription are indicated. The known or predicted functions of the tad locus gene products are indicated. The flp-2 gene, predicted to encode a second pilin, is not expressed or required for pilus biogenesis in A. actinomycetemcomitans. Abbreviation: PP, prepilin peptidase.
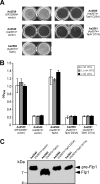
FIG. 2.
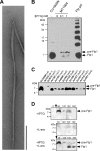
Analysis of Flp1 expression and processing. (A) Transmission electron micrograph of the semipure Flp pilus preparation from A. actinomycetemcomitans CU1000N, stained with uranyl acetate. Bar, 300 nm. (B) Immunoblot analysis of Flp1 pilin expression in A. actinomycetemcomitans wild-type strain CU1000N and E. coli strain BL21 harboring the _tac_p-flp-1 expression construct (strain MT1064) and pilin oligomerization in the semipure Flp pilus preparation. Strain MT1064 was grown in the absence or presence of either 0.1 mM or 1 mM IPTG, as indicated. (C) Flp1 expression and processing in A. actinomycetemcomitans nonpolar tad mutant strains. (D) Processing of pre-Flp1 by TadV in E. coli strain BL21. IPTG and/or
l
-arabinose (L-ara) was added as indicated left of the panel to induce expression of pre-Flp1 or TadV, respectively. Cultures were continuously sampled over a 3-h time course, and the levels and processing state of Flp1 were analyzed by anti-Flp1 immunoblotting. The numbers at the top of the panels indicate time (in minutes) post-IPTG induction. The black arrows at the top of the panels indicate the time of IPTG addition, whereas white arrows indicate the addition of
l
-arabinose. The positions of the pre-Flp1 prepilin and the mature Flp1 pilin are indicated with arrows on the right.
FIG. 3.
(A) Amino acid sequence alignment of A. actinomycetemcomitans TadE and TadF proteins and representative homologs encoded by tad loci in other bacterial species. The processing site of the Flp1 protein of A. actinomycetemcomitans is also aligned. Identical and similar residues are shown in darkly and lightly shaded boxes, respectively. Highly conserved G(−1) and E(+5) residues in the processing sites of the proteins are shown in black boxes. The arrow indicates the processing site. Abbreviations: Aact, A. actinomycetemcomitans; Hduc, H. ducreyi; Pmul, P. multocida; Ypes, Y. pestis. (B) Schematic diagrams of domain organization of the pre-TadF, pre-TadE, and pre-Flp1 proteins of A. actinomycetemcomitans. The signal peptide and hydrophobic domains, as well as portions of the proteins predicted to predominantly have either an α-helical or β-sheet structure, are indicated. The processing sites are indicated with black arrows. (C) Alignment of predicted Shine-Dalgarno sequences upstream of A. actinomycetemcomitans CU1000N flp-1, tadE, and tadF genes. Residues shown in bold are identical to the predicted A. actinomycetemcomitans Shine-Dalgarno sequence. The start codons are shown in bold on a shaded background.
FIG. 4.
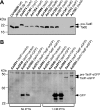
Analysis of expression and processing of the TadE (A) and TadF (B) pseudopilins. (A) Whole-cell extracts of nonpolar tad mutants of A. actinomycetemcomitans were prepared and analyzed by anti-TadE immunoblotting. The positions of pre-TadE and mature TadE protein bands are indicated by black arrows. (B) Analysis of TadF-eGFP expression and processing. The tadF and tadV mutants of A. actinomycetemcomitans harboring either the vector control, a GFP-encoding plasmid, or the TadF-eGFP expression construct were grown in the absence or presence of IPTG, as indicated below the panel. Whole-cell extracts were immunoblotted using the anti-GFP antibody. The black arrows indicate the positions of pre-TadF-eGFP, TadF-eGFP, and GFP.
FIG. 5.
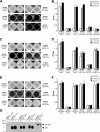
Analysis of Flp1, TadE, and TadF site-directed mutants. Results from qualitative (A, C, and E) and quantitative (B, D, and F) adherence assays examining the effect of G(−1) and E(+5) mutations on Flp1 (A and B), TadE (C and D), and TadF (E and F) functions are shown. The bars indicate standard deviations. (G) Analysis of mutant Flp1 expression and processing. A. actinomycetemcomitans strains were grown in the absence or presence of IPTG, as indicated at the top of the panel. Whole-cell extracts were examined by anti-Flp1 immunoblot analysis. Arrows indicate the positions of the pre-Flp1 and mature Flp1 protein bands.
FIG. 6.
Amino acid alignment of TadV homologs and the predicted topology of the A. actinomycetemcomitans TadV. (A) Identical and similar residues are shown in darkly and lightly shaded boxes, respectively. The highly conserved aspartic acid residues in the predicted catalytic sites are shown in black boxes. The predicted A. actinomycetemcomitans TadV transmembrane domains (TM1 to TM5) are indicated with black lines above the amino acid sequence, and the residues that encompass them are indicated in parentheses. Abbreviations: Aact, A. actinomycetemcomitans; Ccre, C. crescentus; Hduc, H. ducreyi; Pmul, P. multocida; Sfle, Shigella flexneri; Vfis, Vibrio fischeri; Vpar, Vibrio parahaemolyticus; Ypes, Y. pestis, Ypar, Yersinia paratuberculosis. (B) Predicted membrane topologies of A. actinomycetemcomitans TadV and pre-Flp1 proteins. The TadV catalytic aspartic acid residues are shaded dark gray, as are the residues of pre-Flp1, demarcating the prepilin processing site. The site of cleavage of pre-Flp1 is indicated with the black arrow.
FIG. 7.
Functional characterization of the TadV site-directed mutants. (A) Qualitative adherence assay. (B) Quantitative adherence assay. The bars indicate standard deviations. (C) Immunoblot analysis of Flp1 expression and processing. Strains were grown in the presence of 1 mM IPTG. The arrows indicate the positions of the pre-Flp1 and mature Flp1 proteins.
Similar articles
- Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation.
Perez BA, Planet PJ, Kachlany SC, Tomich M, Fine DH, Figurski DH. Perez BA, et al. J Bacteriol. 2006 Sep;188(17):6361-75. doi: 10.1128/JB.00496-06. J Bacteriol. 2006. PMID: 16923904 Free PMC article. - FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili.
de Bentzmann S, Aurouze M, Ball G, Filloux A. de Bentzmann S, et al. J Bacteriol. 2006 Jul;188(13):4851-60. doi: 10.1128/JB.00345-06. J Bacteriol. 2006. PMID: 16788194 Free PMC article. - flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans.
Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. Kachlany SC, et al. Mol Microbiol. 2001 May;40(3):542-54. doi: 10.1046/j.1365-2958.2001.02422.x. Mol Microbiol. 2001. PMID: 11359562 - Posttranslational processing of type IV prepilin and homologs by PilD of Pseudomonas aeruginosa.
Strom MS, Nunn DN, Lory S. Strom MS, et al. Methods Enzymol. 1994;235:527-40. doi: 10.1016/0076-6879(94)35168-6. Methods Enzymol. 1994. PMID: 8057924 Review. - Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa--a review.
Lory S, Strom MS. Lory S, et al. Gene. 1997 Jun 11;192(1):117-21. doi: 10.1016/s0378-1119(96)00830-x. Gene. 1997. PMID: 9224881 Review.
Cited by
- Novel Flp pilus biogenesis-dependent natural transformation.
Angelov A, Bergen P, Nadler F, Hornburg P, Lichev A, Übelacker M, Pachl F, Kuster B, Liebl W. Angelov A, et al. Front Microbiol. 2015 Feb 10;6:84. doi: 10.3389/fmicb.2015.00084. eCollection 2015. Front Microbiol. 2015. PMID: 25713572 Free PMC article. - Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria.
Imam S, Chen Z, Roos DS, Pohlschröder M. Imam S, et al. PLoS One. 2011;6(12):e28919. doi: 10.1371/journal.pone.0028919. Epub 2011 Dec 21. PLoS One. 2011. PMID: 22216142 Free PMC article. - Novel Pelagic Iron-Oxidizing Zetaproteobacteria from the Chesapeake Bay Oxic-Anoxic Transition Zone.
Chiu BK, Kato S, McAllister SM, Field EK, Chan CS. Chiu BK, et al. Front Microbiol. 2017 Jul 18;8:1280. doi: 10.3389/fmicb.2017.01280. eCollection 2017. Front Microbiol. 2017. PMID: 28769885 Free PMC article. - Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon (Salmo salar L.).
Boyd JM, Dacanay A, Knickle LC, Touhami A, Brown LL, Jericho MH, Johnson SC, Reith M. Boyd JM, et al. Infect Immun. 2008 Apr;76(4):1445-55. doi: 10.1128/IAI.01019-07. Epub 2008 Jan 22. Infect Immun. 2008. PMID: 18212071 Free PMC article. - Predictive sequence analysis of the Candidatus Liberibacter asiaticus proteome.
Cong Q, Kinch LN, Kim BH, Grishin NV. Cong Q, et al. PLoS One. 2012;7(7):e41071. doi: 10.1371/journal.pone.0041071. Epub 2012 Jul 18. PLoS One. 2012. PMID: 22815919 Free PMC article.
References
- Akahane, K., D. Sakai, N. Furuya, and T. Komano. 2005. Analysis of the pilU gene for the prepilin peptidase involved in the biogenesis of type IV pili encoded by plasmid R64. Mol. Genet. Genomics 273:350-359. - PubMed
- Armitage, G. C. 1999. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 4:1-6. - PubMed
- Asikainen, S., S. Alaluusua, and L. Saxen. 1991. Recovery of A. actinomycetemcomitans from teeth, tongue, and saliva. J. Periodontol. 62:203-206. - PubMed
- Bardy, S. L., and K. F. Jarrell. 2003. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50:1339-1347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources