Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB - PubMed (original) (raw)
Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB
Zoya M Petrushenko et al. J Biol Chem. 2006.
Abstract
MukBEF is a bacterial SMC (structural maintenance of chromosome) complex required for faithful chromosome segregation in Escherichia coli. The SMC subunit of the complex, MukB, promotes DNA condensation in vitro and in vivo; however, all three subunits are required for the function of MukBEF. We report here that MukEF disrupts MukB x DNA complex. Preassembled MukBEF was inert in DNA binding or reshaping. Similarly, the association of MukEF with DNA-bound MukB served to displace MukB from DNA. When purified from cells, MukBEF existed as a mixture of MukEF-saturated and unsaturated complexes. The holoenzyme was unstable and could only bind DNA upon dissociation of MukEF. The DNA reshaping properties of unsaturated MukBEF were identical to those of MukB. Furthermore, the unsaturated MukBEF was stable and proficient in DNA binding. These results support the view that kleisins are not directly involved in DNA binding but rather bridge distant DNA-bound MukBs.
Figures
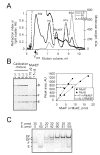
FIGURE 1. Stoichiometry of purified MukEF
A, Light Scattering analysis of MukEF composition. 0.2 mg/ml MukEF was mixed with thyroglobulin (thy; 669 kDa, 8.6 nm), β-amylase (amy; 200 kDa, 4.8 nm) and BSA (66.5 kDa, 3.6 nm) and resolved by gel filtration through a YMC-Pack Diol-300 column. Shown are light scattering (LS) and refraction index (RI) profiles of eluted proteins as well as the calculated molecular weight (MW). Note that the aggregates of thyroglobulin (agg) elute close to the void volume of the column and produce significant LS with little RI. B, Measurement of MukEF stoichiometry from Coomassie Blue staining. Calibration mixture containing 0.55 μM, 2.27 μM and 1.1 μM of individually purified MukB, MukE and MukF, respectively, was electrophoresed next to purified MukEF. Stoichiometry of MukEF was then determined by quantifying the band intensities on Coomassie stained gels. The graph shows band intensities found for the calibration mixture (solid lines) or purified MukEF (dashed lines). C, Reconstitution of MukEF. The indicated amounts of MukE and MukF were incubated in Reconstitution Buffer for 20 min on ice and resolved by electrophoresis through non-denaturing 5% to 20% polyacrylamide gel. The band marked as EF* most likely contains an incompletely assembled MukEF since (i) it is most prominent at low MukE/MukF ratios, and (ii) whenever EF* is present, we also detect the slower complex and free MukF.
FIGURE 2. Bimodal association of MukB and MukEF
A, Sephacryl S300 analysis of reconstituted MukBEF. MukB and MukEF were mixed in indicated proportions and, following reconstitution in 30 μl Reconstitution Buffer, resolved by gel filtration through a 1 ml Sephacryl S300 column equilibrated with the buffer containing 40 mM NaCl, 2 mM MgCl2 (40M), 40 mM NaCl, 2 mM EDTA (40E), or 200 mM NaCl, 2 mM EDTA (200E). MukB and MukBEF migrated with similar mobilities close to the void volume of the column. Eluted proteins were resolved by SDS-PAGE and visualized by silver staining. Positions of MukB (B), MukF (F) and the his-tagged MukE (E9) are indicated on the right. Star marks dimeric MukB that was crosslinked during purification (24). B, Sedimentation analysis of reconstituted MukBEF: Reconstituted MukBEF was mixed with molecular weight standards and resolved by centrifugation through 10% to 40% sucrose gradient in the buffer containing 200 mM NaCl, 2 mM EDTA. The amount of each protein in collected fractions was determined using densitometric analysis of Coomassie stained SDS-PAGE gels. Protein concentrations were further normalized to the highest level found for a given protein across the gradient. Normalized concentrations of MukB, MukF and MukE are plotted against the values of sedimentation coefficient corresponding to each fraction. The E2F-to-B ratio during reconstitution is indicated in the top left corner of each plot. The top panel (0x) shows sedimentation profiles for MukEF and MukB that were analyzed separately from each other. C, Gel filtration of reconstituted MukBEF through Sephacryl S400 in the presence of 200 mM NaCl, 2 mM EDTA. 15 μg MukB was reconstituted with MukEF in indicated proportions in 60 μl and resolved on a 2 ml Sephacryl S400 column. The amount of MukB, MukF and MukE (quantified as for panel B) is plotted against Stokes radii calculated for each fraction from the mobilities of standard proteins. D, Gel filtration of reconstituted MukBEF through Sephacryl S400 in the presence of 40 mM NaCl, 2 mM MgCl2. For the bottom panel, the proteins were eluted at flow rate 0.15 ml/min, which is five-fold faster than in all other experiments.
FIGURE 3. Preassembled MukBEF does not bind or reshape DNA
Following reconstitution, the indicated amounts of MukB and MukEF were assayed for DNA binding and reshaping. Positions of linear (L), nicked circular (NC), supercoiled (SC), relaxed (Rlx) DNA as well as 3-, 4-, 5- and 6-noded knots are indicated on the left of the gels. A, Gel shift analysis of DNA binding by reconstituted MukBEF. 10 ng of pBR322 DNA was incubated with MukBEF and resolved by agarose gel electrophoresis in the presence or absence of 2 mM MgCl2, as indicated. B, Inhibition of DNA supercoiling and knotting by MukEF. DNA knotting (top panel) and supercoiling (bottom panel) reactions were done as described previously (24). M, marker knots generated as previously described (43). C, Inhibition of DNA relaxation by MukB. 10 ng (3.5 fmol) pBR322 DNA was incubated for 10 min with 2.8 pmol of MukB, reconstituted MukBEF, or mock reaction mixture. The reaction mixtures were then treated for 30 min with indicated amounts of wheat germ topoisomerase I, deproteinized, and resolved by agarose gel electrophoresis. D, Inhibition of relaxation by various amounts of MukB. Reactions were done as for panel C.
FIGURE 4
Competition between MukEF and DNA for binding to MukB. A, Time course of DNA displacement by MukEF. 3.5 fmol pBR322 DNA was incubated with 0.7 pmol MukB2 with or without 1 mM MgATP for 30 min. The reactions were then supplemented with mock mixture (0xEF), 0.7 pmol MukE4F2 (1xEF), or 2.8 pmol MukE4F2 (4xEF) and, following indicated times, chilled on ice and analyzed by gel electrophoresis. S, DNA substrate. B, Magnesium induced DNA displacement from BEF. 0.7 pmol MukB2 was reconstituted with 0.7 pmol MukE4F2 and added to 3.5 fmol pBR322 DNA in Reaction Buffer that contained 2 mM EDTA. The reactions were then supplemented with the mock mixture (m) or 4 mM MgCl2 (Mg) and, following indicated times, analyzed by gel electrophoresis (lanes 1–8). For lanes 9–12, magnesium was added to MukBEF before DNA. C, DNA interference with the assembly of MukBEF. 5.8 pmol MukB2 (B) or reconstituted MukB2E4F2 (BEF) was added to Reaction Buffer that contained either 2 mM EDTA (-) or 2 mM MgCl2 (+). At this stage (Stage1), the reaction mixture also contained 5.8 pmol of E4F2 (EF), 1.4 pmol pBR322 DNA (DNA), or mock supplement (m), as indicated. Following 20 min incubation, the reactions were supplemented with 5.8 pmol of E4F2, 1.4 pmol pBR322 DNA, or mock mixture, as indicated (Stage2), further incubated for 5 min, chilled on ice, and resolved by electrophoresis through non-denaturing 4% to 12% gradient gel in TB buffer plus 2 mM MgCl2, for 1 h, 25 V/cm at 4 °C. MukEF, but not MukB or MukBEF can enter the gel under these conditions. Following Coomassie Blue staining, the amount of MukEF was quantified using densitometry. D, Formation of the ternary DNA-MukB-MukEF complex at subsaturating levels of MukEF. 40 pmol MukB2 was reconstituted with 20 pmol MukE4F2, further incubated with (+DNA) or without (-DNA) 2 pmol pBR322 for 10 min in Reaction Buffer containing 2 mM EDTA, and analyzed by gel filtration through Sephacryl S400 equilibrated in the same buffer. Eluted proteins were quantified as described in Fig. 2. To visualize DNA, aliquots of eluted fractions were subjected to agarose gel electrophoresis followed by staining with SYBR Gold. The top panel shows elution profiles for MukB and MukEF analyzed separate from each other.
FIGURE 5. DNA reshaping properties of purified MukBEF
Various topological forms of DNA are the same as described in Fig. 3. A, MukBEF elution profiles after heparin chromatography. Protein concentrations were determined using Bradford assay. MukBEF was purified by nickel-chelate chromatography and further fractionated on a heparin column (Heparin I). The pools of BEF-LS and BEF-HS are marked with the gray and black bars, respectively. BEF-LS was further fractionated by another round of heparin chromatography (Heparin II; the right Y-axis). The fraction numbers for the Heparin II column are indicated beneath the plot. For comparison, the dashed line shows elution of MukB from a Heparin I column. B, Gel shift analysis of DNA binding by BEF-HS and BEF-LS. Reactions were done as described for Fig. 3A. Gel electrophoresis was carried out in TB buffer in the absence or presence (Mg++) of 2 mM MgCl2. C, DNA reshaping activities of BEF-HS and BEF-LS. DNA knotting (top panels) and supercoiling (bottom panels) was analyzed as described in Fig. 3B. Cats, DNA catenanes and oligomers. D, DNA supercoiling activity in Heparin II fractions. BEF-LS was resolved by chromatography through the Heparin II column. The eluted fractions were dialyzed against 20 mM HEPES, pH 7.7, 40 mM NaCl, 8% glycerol, 2 mM EDTA, 1 mM DTT to remove excessive salt and 4 μl aliquots were tested for supercoiling activity. The fraction numbers are the same as in panel A. S, DNA substrate; L, load; FT, flow through. E, Reconstitution of BEF-HS and BEF-LS with MukEF. 40 pmol of BEF-HS or BEF-LS, as indicated, was subjected to reconstitution procedure with 20 pmol of MukEF and resolved by gel-filtration through Sephacryl S300 (see Fig. 2A for details). The columns were equilibrated in the buffer containing 40 mM NaCl, 2 mM MgCl2, with the exception of the third panel, where the column was equilibrated in 200 mM NaCl, 2 mM EDTA (200E). Positions of bands from the molecular weight marker are shown on the left of the top panel. Positions of MukB (B), MukF (F), MukE (E) and the his-tagged MukE (E9) are shown on the right. Note that BEF-HS and BEF-LS contain only the endogenous MukE before reconstitution whereas MukEF contains only the nine-histidine tagged MukE. Thus, the association of MukEF with MukBEF can be followed by evaluating the co-migration of the his-tagged MukE with MukBEF. F, MukBEF fractions eluted from Heparin II differ in their MukEF content. 3 pmol, 6 pmol, 8.5 pmol and 4 pmol MukB2E4F2 from fractions 17, 21, 23 and 26, was reconstituted with 1 pmol, 2 pmol, 2.8 pmol and 1.3 pmol MukE4F2, respectively, and analyzed by gel-filtration as described in panel E.
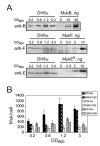
FIGURE 6. MukEF is less abundant in E. coli than MukB
DH5α and MG1655 cells were grown in LB at 37°C, cell aliquots were removed at the indicated turbidities, chilled in ice cold water and pelleted. The pellets were washed and resuspended in 10 mM TrisCl, pH 8.0, 150 mM NaCl, mixed with the SDS-PAGE loading buffer, boiled for 5 min, and 0.1 OD of cells was loaded onto the gel next to the known amounts of purified MukB (top panel) or MukEF (bottom panels) supplemented with cell extract from 0.1 OD of MukBEF-deficient OT7 cells (34). Following gel electrophoresis, MukB, MukF and MukE were transferred onto PVDF membrane and visualized by immuno-staining using appropriate antibody. A, Immunoblot analysis of MukBEF abundance in DH5α cells. B, Copy number of MukB, MukF and MukE in DH5α and MG1655 cells at various cell densities. The molecular mass of MukB, MukF and MukE is, respectively, 170 kDa, 51 kDa and 28 kDa. 1 OD was assumed to contain 109 cells.
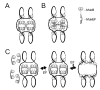
FIGURE 7
Two models of chromatin organization by asymmetric, half-saturated MukBEF. The stoichiometry of active MukBEF is presumed to be B2(E2F). The proteins are postulated to associate further via MukEF-mediated link yielding molecules with the composition B4(E2F)2. The MukEF-saturated complex, B2(E2F)2, does not bind DNA. A, Kleisins directly link distant MukBs, which are bound to separate DNA loops. B, Kleisins promote the attachment of MukB to the chromatin scaffold. C, The relative abundance of MukEF could be varied to control the extent of chromosome compaction. The looped architecture of the chromosome is preserved at limiting levels of MukEF but could be abolished by unbalanced overproduction of the kleisin.
Similar articles
- Biochemical Analysis of Bacterial Condensins.
Petrushenko ZM, Rybenkov VV. Petrushenko ZM, et al. Methods Mol Biol. 2017;1624:145-159. doi: 10.1007/978-1-4939-7098-8_12. Methods Mol Biol. 2017. PMID: 28842882 - MukEF Is required for stable association of MukB with the chromosome.
She W, Wang Q, Mordukhova EA, Rybenkov VV. She W, et al. J Bacteriol. 2007 Oct;189(19):7062-8. doi: 10.1128/JB.00770-07. Epub 2007 Jul 20. J Bacteriol. 2007. PMID: 17644586 Free PMC article. - ATP-induced shrinkage of DNA with MukB protein and the MukBEF complex of Escherichia coli.
Chen N, Zinchenko AA, Yoshikawa Y, Araki S, Adachi S, Yamazoe M, Hiraga S, Yoshikawa K. Chen N, et al. J Bacteriol. 2008 May;190(10):3731-7. doi: 10.1128/JB.01863-07. Epub 2008 Mar 7. J Bacteriol. 2008. PMID: 18326568 Free PMC article. - MukBEF, a chromosomal organizer.
Rybenkov VV, Herrera V, Petrushenko ZM, Zhao H. Rybenkov VV, et al. J Mol Microbiol Biotechnol. 2014;24(5-6):371-83. doi: 10.1159/000369099. Epub 2015 Feb 17. J Mol Microbiol Biotechnol. 2014. PMID: 25732339 Free PMC article. Review. - SMC complexes organize the bacterial chromosome by lengthwise compaction.
Mäkelä J, Sherratt D. Mäkelä J, et al. Curr Genet. 2020 Oct;66(5):895-899. doi: 10.1007/s00294-020-01076-w. Epub 2020 Apr 16. Curr Genet. 2020. PMID: 32300862 Free PMC article. Review.
Cited by
- Physical manipulation of the Escherichia coli chromosome reveals its soft nature.
Pelletier J, Halvorsen K, Ha BY, Paparcone R, Sandler SJ, Woldringh CL, Wong WP, Jun S. Pelletier J, et al. Proc Natl Acad Sci U S A. 2012 Oct 2;109(40):E2649-56. doi: 10.1073/pnas.1208689109. Epub 2012 Sep 14. Proc Natl Acad Sci U S A. 2012. PMID: 22984156 Free PMC article. - The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB.
Li Y, Schoeffler AJ, Berger JM, Oakley MG. Li Y, et al. J Mol Biol. 2010 Jan 8;395(1):11-9. doi: 10.1016/j.jmb.2009.10.040. Epub 2009 Oct 22. J Mol Biol. 2010. PMID: 19853611 Free PMC article. - A folded conformation of MukBEF and cohesin.
Bürmann F, Lee BG, Than T, Sinn L, O'Reilly FJ, Yatskevich S, Rappsilber J, Hu B, Nasmyth K, Löwe J. Bürmann F, et al. Nat Struct Mol Biol. 2019 Mar;26(3):227-236. doi: 10.1038/s41594-019-0196-z. Epub 2019 Mar 4. Nat Struct Mol Biol. 2019. PMID: 30833788 Free PMC article. - Kinetic control of TolC recruitment by multidrug efflux complexes.
Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. Tikhonova EB, et al. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16416-21. doi: 10.1073/pnas.0906601106. Epub 2009 Sep 2. Proc Natl Acad Sci U S A. 2009. PMID: 19805313 Free PMC article. - The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV.
Hayama R, Bahng S, Karasu ME, Marians KJ. Hayama R, et al. J Biol Chem. 2013 Mar 15;288(11):7653-7661. doi: 10.1074/jbc.M112.418087. Epub 2013 Jan 24. J Biol Chem. 2013. PMID: 23349462 Free PMC article.
References
- Swedlow JR, Hirano T. Mol Cell. 2003;11:557–569. - PubMed
- Cobbe N, Heck MM. J Struct Biol. 2000;129:123–143. - PubMed
- Nasmyth K, Haering CH. Annu Rev Biochem. 2005;74:595–648. - PubMed
- Koshland D, Strunnikov A. Annu Rev Cell Dev Biol. 1996;12:305–333. - PubMed
- Hiraga S. Annu Rev Genet. 2000;34:21–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases