Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) - PubMed (original) (raw)
. 2006 Oct 1;20(19):2660-72.
doi: 10.1101/gad.397006. Epub 2006 Sep 18.
Affiliations
- PMID: 16983144
- PMCID: PMC1578693
- DOI: 10.1101/gad.397006
Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS)
Katja Vanselow et al. Genes Dev. 2006.
Abstract
PERIOD (PER) proteins are central components within the mammalian circadian oscillator, and are believed to form a negative feedback complex that inhibits their own transcription at a particular circadian phase. Phosphorylation of PER proteins regulates their stability as well as their subcellular localization. In a systematic screen, we have identified 21 phosphorylated residues of mPER2 including Ser 659, which is mutated in patients suffering from familial advanced sleep phase syndrome (FASPS). When expressing FASPS-mutated mPER2 in oscillating fibroblasts, we can phenocopy the short period and advanced phase of FASPS patients' behavior. We show that phosphorylation at Ser 659 results in nuclear retention and stabilization of mPER2, whereas phosphorylation at other sites leads to mPER2 degradation. To conceptualize our findings, we use mathematical modeling and predict that differential PER phosphorylation events can result in opposite period phenotypes. Indeed, interference with specific aspects of mPER2 phosphorylation leads to either short or long periods in oscillating fibroblasts. This concept explains not only the FASPS phenotype, but also the effect of the tau mutation in hamster as well as the doubletime mutants (dbtS and dbtL ) in Drosophila.
Figures
Figure 8.
Schematic representation of PER2 phosphorylation sites and their functional impact on the circadian oscillator. (A) Linear map of the PER2 protein with positions of phosphorylated residues identified by mass spectrometry and additional functionally important domains (see also Table 1). (Blue lines) Sites phosphorylated by endogenous kinases in cells; (red asterisks) sites also phosphorylated in vitro by CKIδ. Functional domains: (PAS) PER-ARNT-SIM domain; (NES) nuclear export sequence; (NLS) nuclear localization signal; (CKIε) CKIε-binding domain; (CRY) CRY1/2-binding domain. (B) Model for the differential effects of PER2 phosphorylation on circadian oscillations. PER2 contains at least two functionally different sets of phosphorylation sites—one primarily mediating proteasomal degradation (green), the other nuclear retention (purple). In FASPS-PER2 (right side of the panels), the latter cannot be phosphorylated because Ser 659 is mutated to glycine (Toh et al. 2001). At the beginning of the circadian cycle (morning/ midday), newly synthesized PER2 protein shuttles between nucleus and cytoplasm, where it is phosphorylated by kinases such as CKIε/δ at sites that target it for rapid proteasomal degradation in the cytoplasm. Later (afternoon/early night), complex formation with CRY proteins enhances the nuclear localization of the PER2–CRY complex and likely activates or recruits additional kinases to the PER2–CRY complex. These yet-unknown kinases phosphorylate PER2 at the FASPS site, which serves as a priming site for CKIε/δ phosphorylation at downstream residues. Together, this leads to nuclear accumulation of the PER2–CRY complex and thereby to transcriptional repression of CLOCK–BMAL1 transactivation. At the end of the circadian cycle (late night), the PER2–CRY repression is released because the PER2–CRY complex is degraded in the cytoplasm after nuclear export. In FASPS-PER2, however, the region responsible for nuclear retention cannot be phosphorylated (red crosses), leading to premature nuclear export of the PER2–CRY complex, and thus to an earlier cytosolic degradation and to a faster circadian cycle.
Figure 1.
Mouse PER2 protein is phosphorylated at a serine residue, which causes FASPS in humans. (A) The alignment of the amino acid sequence of human and mouse PER2 shows a highly conserved cluster of serine and threonine residues in the region where the FASPS mutation has been identified. In patients suffering from FASPS, one copy of hPER2 is mutated at position 662 (serine to glycine). Those phosphorylatable residues—which are identical in PER1, PER2, and PER3—are bold and underlined. (B) The FASPS site of mPER2 is phosphorylated in tissue culture cells. Whole-cell lysate from HEK293 cells stably expressing V5-epitope-tagged mPER2 was used to immunoprecipitate mPER2. After separation by gel electrophoresis, mPER2 protein was digested with proteases, and the resulting phosphopeptides were enriched using titansphere chromatography and subsequently identified using nano-liquid chromatography followed by tandem mass spectrometry (nanoLC-MS/ MS), as described (Schlosser et al. 2005). (C) PCR-generated mutations were made in the conserved serine–threonine cluster of mPER2 for subsequent comparison with wild type in various assays.
Figure 2.
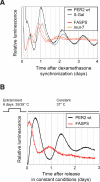
The FASPS mutation leads to an advanced phase and a shorter period in oscillating fibroblasts. (A) Real-time circadian oscillations of luciferase reporter activity in dexamethasone-synchronized NIH3T3 fibroblasts, which stably express either PER2 wild-type or the PER2 mutants FASPS and mut-7 as well as β-galactosidase as a control from the same chromosomal locus (see main text and Materials and Methods). Shown are representative detrended time series from at least five independent experiments. (B) PER2 wild-type and FASPS-expressing NIH3T3 cell lines were entrained with temperature cycles consisting of 12 h at 35°C, and 12 h at 39°C for 6 d, as indicated. After transfer to constant 37°C, real-time circadian oscillations of luciferase reporter activity were recorded. Shown are representative detrended time series from at least three independent experiments.
Figure 3.
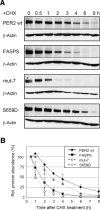
The FASPS mutation leads to a destabilization of the PER2 protein. (A) NIH3T3 cells stably expressing either PER2 wild-type or the indicated PER2 mutant proteins from the same chromosomal locus were treated with the protein translation inhibitor CHX. Cells were harvested at the indicated times after CHX treatment, and the amount of PER2 protein was determined by Western blotting. Treatment of the cells with solvent did not lead to a decrease of protein amount (not shown). (B) Quantification of the PER2 protein from the experiments described above. Error bars represent the range from two independent experiments.
Figure 4.
The FASPS mutation leads to an increased sensitivity of PER2 toward CKIε-mediated degradation. (A) HEK293 cells coexpressing the indicated PER2 variants and either CKIε(wt) or the kinase-dead variant CKIε(K38A) were analyzed for PER2 protein abundance by Western blotting. Plasmid expressing β-galactosidase was cotransfected for normalization. (B) Quantification of the experiments described above with β-galactosidase band intensity used for normalization. The ratio between PER2 protein abundance when coexpressed with CKIε(wt) and PER2 protein abundance without coexpression of the kinase (average ± SEM, n = 2–5) is plotted. For FASPS and mut-7, the coexpression of CKIε leads to a significantly larger decrease in protein abundance than for PER2 wild type ([*] p < 0.05; [**] p < 0.01; unpaired homoscedastic _t_-test). (C) HEK293 cells expressing the indicated PER2 variants with or without CRY1 were analyzed for PER2 protein abundance by Western blotting. (D) HEK293 cells coexpressing CRY1 and either PER2 wild type or mut-7 were treated with the specific casein kinase I inhibitor CKI-7 or solvent. The effect on PER2 protein abundance was analyzed by Western blotting.
Figure 5.
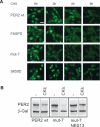
The FASPS mutation leads to a premature nuclear clearance. (A) PER2 variant-expressing NIH3T3 cell lines were treated with the protein translation inhibitor CHX. Cells were fixed at the indicated time points after treatment and immunostained with anti-V5 and fluorescently labeled secondary antibody. Shown are representative fields of two experiments. Bar, 20 μm. (B) HEK293 cells were transiently transfected with either PER2 wild type, mut-7, or a mut-7 variant, whose nuclear localization signals 1 and 3 (mut-7 NES13) were mutated. CKIε was coexpressed where indicated, and PER2 protein abundance was detected by Western blotting.
Figure 6.
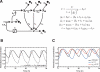
Mathematical model of PER2 phosphorylation. (A) Scheme and differential equations of our conceptual model of the circadian oscillator with emphasis on phosphorylation events. The variables y 0, y 1, and y 12 represent the different phosphorylated protein species with y 0 being nonphosphorylated protein, y 1 phosphorylated at “one” site, and y 12 phosphorylated at “two” sites. The variables x and z describe mRNA and nuclear protein, respectively. (Black arrows) Net fluxes of the different species; (open arrow) translation; (chopped arrow with box) transcriptional inhibition using a Hill term; (black dots) degradation. The differential equations are chosen as simply as possible and are essentially an extension of the Goodwin oscillator model (Goodwin 1965). For a full reasoning of the model design and parameter choice, see main text and Supplemental Material. (B) With the parameters chosen, the model shows sustained 24-h oscillations with the mRNA peak preceding the peak of total protein by ~6 h, thus reproducing experimental results. (C) Interference with the phosphorylation on the “second” site, as is the case in FASPS, is modeled by reducing the respective phosphorylation rate constant parameter q 12, leading to a shorter circadian period (red) compared with a wild-type situation (black). In contrast, if phosphorylation of “both” sites is blocked (modeled by reducing q 1 to 0), it is predicted that the circadian period lengthens.
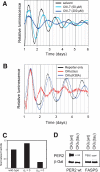
Figure 7.
Experimental test of the model's predictions. (A) Dexamethasone-synchronized NIH3T3 reporter cells were treated with the specific CKI-inhibitor CKI-7 in different concentrations or with solvent (DMSO) only. Shown are representative detrended time series from at least five independent experiments, which were aligned to the first peak for period comparison. (B) NIH3T3 cells were stably transfected with circadian luciferase reporter and either CKIε(tau) or the kinase-dead version CKIε(K38A) and synchronized with dexamethasone. Shown are representative detrended and normalized time series from two independent experiments, which were aligned to the first peak for period comparison. (C) Prediction of the model for the effect of CKIε(tau) on PER2 protein half-life. The reduced kinase activity of CKIε(tau) in vitro is modeled by either reducing the “first” phosphorylation (q 1 = 0), the “second” phosphorylation (q 12 = 0), or “both” (q 1 = 0). Since only the reduction of the “second” phosphorylation event (q 12 = 0) results in a short circadian period as seen in hamsters, the model predicts a destabilization of PER2 by CKIε(tau) in comparison with CKIε(wt). For the calculation of half-lives, see Supplemental Material. (D) Experiments verify the model's predictions: CKIε(wt) or CKIε(tau) was coexpressed with wild-type or FASPS PER2, and steady-state protein abundance levels were analyzed in Western blot experiments. CKIε(tau) destabilizes mPER2 to a much higher extent than CKIε(wt).
Comment in
- The right place at the right time: regulation of daily timing by phosphorylation.
Merrow M, Mazzotta G, Chen Z, Roenneberg T. Merrow M, et al. Genes Dev. 2006 Oct 1;20(19):2629-3. doi: 10.1101/gad.1479706. Genes Dev. 2006. PMID: 17015426 Review. No abstract available.
Similar articles
- Role of phosphorylation in the mammalian circadian clock.
Vanselow K, Kramer A. Vanselow K, et al. Cold Spring Harb Symp Quant Biol. 2007;72:167-76. doi: 10.1101/sqb.2007.72.036. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419274 Review. - Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics.
Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. Reischl S, et al. J Biol Rhythms. 2007 Oct;22(5):375-86. doi: 10.1177/0748730407303926. J Biol Rhythms. 2007. PMID: 17876059 - Modeling of a human circadian mutation yields insights into clock regulation by PER2.
Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptácek LJ. Xu Y, et al. Cell. 2007 Jan 12;128(1):59-70. doi: 10.1016/j.cell.2006.11.043. Cell. 2007. PMID: 17218255 Free PMC article. - Casein kinase 1-dependent phosphorylation of familial advanced sleep phase syndrome-associated residues controls PERIOD 2 stability.
Shanware NP, Hutchinson JA, Kim SH, Zhan L, Bowler MJ, Tibbetts RS. Shanware NP, et al. J Biol Chem. 2011 Apr 8;286(14):12766-74. doi: 10.1074/jbc.M111.224014. Epub 2011 Feb 15. J Biol Chem. 2011. PMID: 21324900 Free PMC article. - Novel insights from genetic and molecular characterization of the human clock.
Ptácek LJ, Jones CR, Fu YH. Ptácek LJ, et al. Cold Spring Harb Symp Quant Biol. 2007;72:273-7. doi: 10.1101/sqb.2007.72.017. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419283 Review.
Cited by
- Period 2 Regulates CYP2B10 Expression and Activity in Mouse Liver.
Chen M, Chen M, Lu D, Wang Y, Zhang L, Wang Z, Wu B. Chen M, et al. Front Pharmacol. 2021 Nov 23;12:764124. doi: 10.3389/fphar.2021.764124. eCollection 2021. Front Pharmacol. 2021. PMID: 34887762 Free PMC article. - Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock.
St John PC, Hirota T, Kay SA, Doyle FJ 3rd. St John PC, et al. Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):2040-5. doi: 10.1073/pnas.1323618111. Epub 2014 Jan 21. Proc Natl Acad Sci U S A. 2014. PMID: 24449901 Free PMC article. - A Robust Model for Circadian Redox Oscillations.
Del Olmo M, Kramer A, Herzel H. Del Olmo M, et al. Int J Mol Sci. 2019 May 13;20(9):2368. doi: 10.3390/ijms20092368. Int J Mol Sci. 2019. PMID: 31086108 Free PMC article. - A role for timely nuclear translocation of clock repressor proteins in setting circadian clock speed.
Lee E, Kim EY. Lee E, et al. Exp Neurobiol. 2014 Sep;23(3):191-9. doi: 10.5607/en.2014.23.3.191. Epub 2014 Sep 18. Exp Neurobiol. 2014. PMID: 25258565 Free PMC article. Review. - The Function, Regulation, and Mechanism of Protein Turnover in Circadian Systems in Neurospora and Other Species.
Zhang H, Zhou Z, Guo J. Zhang H, et al. Int J Mol Sci. 2024 Feb 22;25(5):2574. doi: 10.3390/ijms25052574. Int J Mol Sci. 2024. PMID: 38473819 Free PMC article. Review.
References
- Balsalobre, A., Damiola, F., Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. - PubMed
- Brown, S.A., Zumbrunn, G., Fleury-Olela, F., Preitner, N., Schibler, U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 2002;12:1574–1583. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases