PPAR{alpha} mediates the hypolipidemic action of fibrates by antagonizing FoxO1 - PubMed (original) (raw)
PPAR{alpha} mediates the hypolipidemic action of fibrates by antagonizing FoxO1
Shen Qu et al. Am J Physiol Endocrinol Metab. 2007 Feb.
Abstract
High-fructose consumption is associated with insulin resistance and diabetic dyslipidemia, but the underlying mechanism is unclear. We show in hamsters that high-fructose feeding stimulated forkhead box O1 (FoxO1) production and promoted its nuclear redistribution in liver, correlating with augmented apolipoprotein C-III (apoC-III) production and impaired triglyceride metabolism. High-fructose feeding upregulated peroxisome proliferator-activated receptor-gamma coactivator-1beta and sterol regulatory element binding protein-1c expression, accounting for increased fat infiltration in liver. High-fructose-fed hamsters developed hypertriglyceridemia, accompanied by hyperinsulinemia and glucose intolerance. These metabolic aberrations were reversible by fenofibrate, a commonly used anti-hypertriglyceridemia agent that is known to bind and activate peroxisome proliferator-activated receptor-alpha (PPARalpha). PPARalpha physically interacted with, but functionally antagonized, FoxO1 in hepatic apoC-III expression. These data underscore the importance of FoxO1 deregulation in the pathogenesis of hypertriglyceridemia in high-fructose-fed hamsters. Counterregulation of hepatic FoxO1 activity by PPARalpha constitutes an important mechanism by which fibrates act to curb apoC-III overproduction and ameliorate hypertriglyceridemia.
Figures
Fig. 1
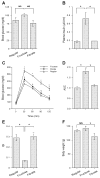
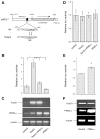
Effect of fenofibrate on glucose metabolism in high-fructose-fed hamsters. A: blood glucose levels. B: plasma insulin levels. C: intraperitoneal glucose tolerance. D: area under the curve (AUC) of blood glucose profiles in response to glucose tolerance. After normalizing to regular chow-treated hamsters, the relative AUC values were plotted. E: insulin sensitivity indexes (ISI). F: body weight. Blood glucose and plasma insulin levels were determined after 16-h fasting. Data were obtained from hamsters fed on regular chow (regular, n = 8), high-fructose diet (fructose, n = 9), and high-fructose plus fenofibrate treatment (fibrate, n = 9) after 4-wk treatment with fenofibrate. NS, not significant. *P < 0.05 by ANOVA.
Fig. 2
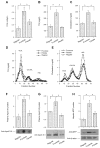
Effect of fenofibrate on plasma lipid metabolism. A: plasma free fatty acid (FFA) levels. B: plasma triglyceride (TG) levels. C: plasma cholesterol levels. Data were obtained following 4-wk treatment with fenofibrate. Aliquots (500 μl) of plasma pooled from killed hamsters in individual groups were subjected to gel filtration column chromatography through 2 consecutive Tricom high-performance Superose S-6 10/300GL columns in a fast protein liquid chromatography (FPLC) system. Seventy fractions were collected for the determination of TG (D) and cholesterol (E) concentrations. Plasma apolipoprotein C-III (apoC-III) levels were determined by dot blot (F) and Western blot (G) assays. Dot blot assay seemed to result in an overestimation of plasma apoC-III levels. H: hepatic microsomal TG transfer protein (MTP) levels were determined by semiquantitative Western blot assay. LDL, lipoprotein lipase; IDL, intermediate-density lipoprotein; HDL, high-density lipoprotein; VLDL, very-low-density lipoprotein. *P < 0.05.
Fig. 3
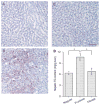
Hepatic lipid content. Hamsters were killed after 4 wk of fibrate treatment. Liver tissues of hamsters treated with regular chow (A), high-fructose diet (B), and high-fructose diet plus fenofibrate (C) were embedded with Histoprep tissue embedding media. Frozen sections (8 μm) were cut and stained with Oil red O, followed by counterstaining with hematoxylin. D: hepatic lipid content in different groups of hamsters was also determined using a quantitative hepatic TG assay. Bar = 50 μm. *P < 0.001.
Fig. 4
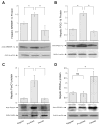
Hepatic gene expression. Sterol regulatory element binding protein-1c (SREBP-1c; A), peroxisome proliferator-activated receptor-γ coactivator-1β (PGC-1β; B), forkhead box O1 (FoxO1; C), and peroxisome proliferator-activated receptor-α (PPARα; D) protein levels in livers of euthanized hamsters were determined by semiquantitative immunoblot assays. *P < 0.05.
Fig. 5
FoxO1 immunohistochemistry. Liver tissue of hamsters fed regular chow (A, B, and C), high-fructose diet (D, E, and F), and high-fructose diet plus fenofibrate treatment (G, H, and I) was embedded with the Histoprep tissue embedding media. Frozen sections (8 μm) were cut and incubated with rabbit anti-FoxO1 antibody (A, D, and G), followed by immunostaining with Cy3-conjugated goat anti-rabbit IgG (red). The nuclei of hepatocytes (B, E, and H) were stained with the TO-PRO-3 dye (blue). Bar =10 μm.
Fig. 6
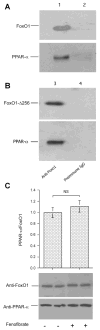
FoxO1 and PPARα interaction. A: immunoblots. Liver tissue (20 mg) of fenofibrate-treated high-fructose-fed hamsters was homogenized and subjected to immunoprecipitation using rabbit anti-FoxO1 antibody (lane 1) or rabbit preimmune serum (lane 2). Immunoprecipitates were analyzed by immunoblot assay using anti-FoxO1 and anti-PPARα antibodies, respectively. B: immunoblots. HepG2 cells were transfected with pPPARα and transduced with adenoviral vector expressing FoxO1-Δ256. After 48-h incubation, cells were subjected to immunoprecipitation by rabbit anti-FoxO1 antibody (lane 3) or preimmune serum (lane 4), followed by immunoblot analysis using anti-FoxO1 or anti-PPARα antibody. C: effect of fenofibrate on FoxO1 and PPARα interaction. HepG2 cells expressing FoxO1 and PPARα were cultured in the presence and absence of 50 mM fenofibrate for 16 h, followed by immunoprecipitation using anti-FoxO1 antibody. Immunoprecipitated proteins were subjected to semiquantitative immunoblot assay. The amounts of PPARα were normalized to FoxO1 and compared between mock and fenofibrate treatment groups. Data were from 3 independent experiments.
Fig. 7
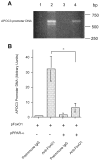
PPARα counteracts FoxO1 in apoC-III expression. A: the human APOC3 promoter-directed luciferase system in pHD317. The nucleotide sequences corresponding to the insulin-responsive element (IRE) in pHD317 and its mutant version in pHD334 were underlined. B: APOC3 promoter activity. HepG2 cells were transfected with pHD317 together with plasmids expressing FoxO1 or PPARα alone or both in combination, using pCA35-LacZ as control for the normalization of transfection efficiency. After 48-h incubation, cells were collected for the determination of luciferase and β-galactosidase activities. The ratios between luciferase and β-galactosidase activities were compared between different conditions. Results were obtained from 5 independent experiments. *P < 0.001. C: RT-PCR products of FoxO1, PPARα, and β-actin mRNA. A subset of HepG2 cells treated identically as described in B was used for the preparation of total RNA, which was analyzed by RT-PCR using specific primers corresponding to FoxO1, PPARα, and β-actin cDNA. The RT-PCR products were resolved on 1.0% agarose gels and visualized under UV light after staining with ethidium bromide. D: the mutant APOC3 promoter activity. HepG2 cells were transfected with pHD334 expressing the mutant APOC3 promoter-directed luciferase expression system in the presence of FoxO1 or PPARα alone or both in combination, using pCA35-LacZ as control. Each condition was run in triplicate. Cells were subjected to luciferase and β-galactosidase activity assay after 24-h incubation. E: effect of PPARα on FoxO1 promoter activity. The FoxO1 promoter-directed luciferase expression system pFoxO-luc was cotransfected with pPPARα or control pGL3-basic plasmid (2 μg per plasmid) in 6-well plates using pCA35-LacZ (1 μg) as control. After 24-h incubation, cells were collected for the determination of luciferase and β-galactosidase activities. Data were from 3 independent experiments. #P < 0.05. F: effect of PPARα on FoxO1 mRNA expression. A subset of HepG2 cells from experiment in E was used for the preparation of total RNA, which was analyzed by RT-PCR using specific primers corresponding to FoxO1, PPARα, and β-actin cDNA.
Fig. 8
PPARα interferes with FoxO1 binding to apoC-III promoter. A: chromatin immunoprecipitation (ChIP) analysis of FoxO1-APOC3 promoter DNA interaction in the absence and presence of PPARα production. HepG2 cells were transfected with pFoxO1 alone (lanes 1 and 2) or cotransfected with both pFoxO1 and pPPARα plasmids (lanes 3 and 4) in 6-well dishes. After 48-h incubation, cells were subjected to ChIP assay using preimmune IgG (lanes 1 and 3) or rabbit anti-FoxO1 antibody (lanes 2 and 4). Immunoprecipitates were analyzed by PCR using 20-nucleotide (nt) primers flanking −675/+1 nt region of human APOC3 promoter DNA. The PCR products were analyzed on 1% agarose gels and visualized under UV light after ethidium bromide staining. B: quantification of PCR products. The relative amounts of PCR products corresponding to the specific APOC3 promoter DNA were quantified by densitometry. Results were obtained from 3 independent experiments. *P < 0.001.
Similar articles
- Fenofibrate lowers lipid accumulation in myotubes by modulating the PPARα/AMPK/FoxO1/ATGL pathway.
Chen WL, Chen YL, Chiang YM, Wang SG, Lee HM. Chen WL, et al. Biochem Pharmacol. 2012 Aug 15;84(4):522-31. doi: 10.1016/j.bcp.2012.05.022. Epub 2012 Jun 9. Biochem Pharmacol. 2012. PMID: 22687626 - Peroxisome proliferator-activated receptor α activation induces hepatic steatosis, suggesting an adverse effect.
Yan F, Wang Q, Xu C, Cao M, Zhou X, Wang T, Yu C, Jing F, Chen W, Gao L, Zhao J. Yan F, et al. PLoS One. 2014 Jun 13;9(6):e99245. doi: 10.1371/journal.pone.0099245. eCollection 2014. PLoS One. 2014. PMID: 24926685 Free PMC article. - Activation of PPAR alpha by fenofibrate inhibits apoptosis in vascular adventitial fibroblasts partly through SIRT1-mediated deacetylation of FoxO1.
Wang WR, Liu EQ, Zhang JY, Li YX, Yang XF, He YH, Zhang W, Jing T, Lin R. Wang WR, et al. Exp Cell Res. 2015 Oct 15;338(1):54-63. doi: 10.1016/j.yexcr.2015.07.027. Epub 2015 Jul 28. Exp Cell Res. 2015. PMID: 26226216 - Targeting FoxO1 for hypertriglyceridemia.
Kim DH, Zhang T, Ringquist S, Dong HH. Kim DH, et al. Curr Drug Targets. 2011 Aug;12(9):1245-55. doi: 10.2174/138945011796150262. Curr Drug Targets. 2011. PMID: 21443465 Review. - Mode of action of fibrates in the regulation of triglyceride and HDL-cholesterol metabolism.
Fruchart JC, Duriez P. Fruchart JC, et al. Drugs Today (Barc). 2006 Jan;42(1):39-64. doi: 10.1358/dot.2006.42.1.963528. Drugs Today (Barc). 2006. PMID: 16511610 Review.
Cited by
- The transcription factors CREBH, PPARa, and FOXO1 as critical hepatic mediators of diet-induced metabolic dysregulation.
Yang Z, Roth K, Agarwal M, Liu W, Petriello MC. Yang Z, et al. J Nutr Biochem. 2021 Sep;95:108633. doi: 10.1016/j.jnutbio.2021.108633. Epub 2021 Mar 28. J Nutr Biochem. 2021. PMID: 33789150 Free PMC article. Review. - L-carnitine attenuates autophagic flux, apoptosis, and necroptosis in rats with dexamethasone-induced non-alcoholic steatohepatitis.
Amer AE, Ghoneim HA, Abdelaziz RR, Shehatou GSG, Suddek GM. Amer AE, et al. BMC Pharmacol Toxicol. 2024 Dec 30;25(1):102. doi: 10.1186/s40360-024-00820-z. BMC Pharmacol Toxicol. 2024. PMID: 39736705 Free PMC article. - FAM3B (PANDER) functions as a co-activator of FOXO1 to promote gluconeogenesis in hepatocytes.
Chi Y, Meng Y, Wang J, Yang W, Wu Z, Li M, Wang D, Gao F, Geng B, Tie L, Zhang W, Yang J. Chi Y, et al. J Cell Mol Med. 2019 Mar;23(3):1746-1758. doi: 10.1111/jcmm.14073. Epub 2018 Nov 28. J Cell Mol Med. 2019. PMID: 30488666 Free PMC article. - Down-regulation of MicroRNA-592 in obesity contributes to hyperglycemia and insulin resistance.
Song Y, Wu L, Li M, Xiong X, Fang Z, Zhou J, Yan G, Chen X, Yang J, Li Y. Song Y, et al. EBioMedicine. 2019 Apr;42:494-503. doi: 10.1016/j.ebiom.2019.03.041. Epub 2019 Apr 1. EBioMedicine. 2019. PMID: 30948354 Free PMC article. - Chronic activation of PPARα with fenofibrate reduces autophagic proteins in the liver of mice independent of FGF21.
Jo E, Li S, Liang Q, Zhang X, Wang H, Herbert TP, Jenkins TA, Xu A, Ye JM. Jo E, et al. PLoS One. 2017 Apr 19;12(4):e0173676. doi: 10.1371/journal.pone.0173676. eCollection 2017. PLoS One. 2017. PMID: 28422956 Free PMC article.
References
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. - PubMed
- Altomonte J, Richter A, Harbaran S, Suriawinata j, Nakae J, Thung SN, Meseck M, Accili D, Dong H. Inhibition of Foxo1 function is associated with improved fasting glycemia in diabetic mice. Am J Physiol Endocrinol Metab. 2003;285:E718–E728. - PubMed
- Avramoglu RK, Qiu W, Adeli K. Mechanisms of metabolic dyslipidemia in insulin resistant states: deregulation of hepatic and intestinal lipoprotein secretion. Front Biosci. 2003;8:d464–d476. - PubMed
- Bard JM, Charles MA, Juhan-Vague I, Vague P, Andre P, Safar M, Fruchart JC, Eschwege E. Accumulation of triglyceride-rich lipoprotein in subjects with abdominal obesity. Arterioscler Thromb Vase Biol. 2001;21:407–414. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous