Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes - PubMed (original) (raw)
doi: 10.1186/gb-2006-7-9-r84.
Jo Vandesompele, Pierre Heimann, Nurten Yigit, Siv Beckman, Alexander Schramm, Angelika Eggert, Raymond L Stallings, Yves Benoit, Marleen Renard, Anne De Paepe, Geneviève Laureys, Sven Påhlman, Frank Speleman
Affiliations
- PMID: 16989664
- PMCID: PMC1794547
- DOI: 10.1186/gb-2006-7-9-r84
Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes
Katleen De Preter et al. Genome Biol. 2006.
Erratum in
- Genome Biol. 2007;8(1):401
Abstract
Background: Neuroblastoma tumor cells are assumed to originate from primitive neuroblasts giving rise to the sympathetic nervous system. Because these precursor cells are not detectable in postnatal life, their transcription profile has remained inaccessible for comparative data mining strategies in neuroblastoma. This study provides the first genome-wide mRNA expression profile of these human fetal sympathetic neuroblasts. To this purpose, small islets of normal neuroblasts were isolated by laser microdissection from human fetal adrenal glands.
Results: Expression of catecholamine metabolism genes, and neuronal and neuroendocrine markers in the neuroblasts indicated that the proper cells were microdissected. The similarities in expression profile between normal neuroblasts and malignant neuroblastomas provided strong evidence for the neuroblast origin hypothesis of neuroblastoma. Next, supervised feature selection was used to identify the genes that are differentially expressed in normal neuroblasts versus neuroblastoma tumors. This approach efficiently sifted out genes previously reported in neuroblastoma expression profiling studies; most importantly, it also highlighted a series of genes and pathways previously not mentioned in neuroblastoma biology but that were assumed to be involved in neuroblastoma pathogenesis.
Conclusion: This unique dataset adds power to ongoing and future gene expression studies in neuroblastoma and will facilitate the identification of molecular targets for novel therapies. In addition, this neuroblast transcriptome resource could prove useful for the further study of human sympathoadrenal biogenesis.
Figures
Figure 1
Identification of sympathetic neuroblasts and chromaffin cells in human fetal adrenal glands by immunohistochemical analysis. Sections of a human fetal (19 weeks) adrenal gland, adjacent to those used for laser capture retrieval of cells for mRNA extraction and gene expression profiling, were stained with (a) hematoxylin and eosin or antibodies directed against (b,f) TH, (c,g) CHGA, (d,h) BCL2, and (e,i) HNK1. Whereas the immunoreactivities of BCL2 and HNK1 are specific for neuroblasts, TH and CHGA expression is pronounced in chromaffin cells and weak in neuroblasts [8]. Stars indicate chromaffin cells (TH+, CHGA+, BCL2-, and HNK1-), either solitary or intermingled with neuroblasts. Panels a-e show a cluster of adrenal neuroblasts and panels f-i show cortical area within scattered chromaffin cells adjacent to the neuroblast cluster. Inserts in panels b-e (bars: 10 μm) correspond to the boxed areas in these panels (bars in panels a-i: 100 μm). BCL2, B-cell CLL/lymphoma 2; CHGA, chromogranin A; H&E, hematoxylin and eosin; HNK1, carbohydrate epitope; TH, tyrosine hydroxylase.
Figure 2
Laser capture microdissection of neuroblast clusters. (a) Large cluster of neuroblasts in fetal adrenal glands at 19 weeks' gestational age (mounted hematoxylin and eosin stained cryosections), (b,c) unmounted hematoxylin and eosin stained fetal adrenal cryosections with a neuroblast cluster before and after microdissection (sample 2), and (d) the microdissected neuroblast cluster.
Figure 3
Multidimensional scaling of neuroblast, cortex, and neuroblastoma samples. (a) Multidimensional scaling of neuroblast, cortex, and neuroblastoma samples using all genes (Spearman correlation) and (b) multidimensional scaling of neuroblast, cortex, neuroblastoma, 79 normal tissue samples and other cancer samples (in duplo), and three neural stem cell cultures using the genes that are differentially expressed between fetal adrenal neuroblast and fetal adrenal cortex shows that the neuroblasts cluster very close to the neuroblastomas.
Figure 4
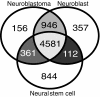
Venn diagram analysis of the genes with detectable expression in neuroblastoma, neuroblast, and cortex samples. (a) All genes, (b) transcription factors (GO:0003700), and (c) neurogenesis genes (GO:0007399). The number of genes that are in common between neuroblast and neuroblastoma is higher than the number of genes that are in common between the neuroblasts and cortex samples (especially for the gene classes transcription and neurogenesis), indicating that neuroblastomas resemble neuroblasts. GO, Gene Ontology.
Figure 5
Positional gene enrichment analysis of genes on chromosome 17. Positional gene enrichment analysis for the genes that are more highly expressed in neuroblastoma compared to normal neuroblasts identified two regions on 17q with significant over-representation (-10log P values; indicated in grey; the genes in these regions are printed in the boxes). The horizontal red line indicates the multiple testing corrected P value of 0.01, above which the positional gene enrichment value denotes significant over-representation. Vertical lines show the position of the genes on chromosome 17 from the gene list under investigation. The boxplot shows the gene density along the chromosome.
Figure 6
Venn diagram analysis of genes with detectable expression in neuroblast, neuroblastoma, and neural stem cell lines. This analysis shows that neuroblasts have many genes in common with neuroblastoma, but it also demonstrates that neural stem cell lines have more genes in common with the neuroblastomas than with the normal neuroblasts.
Similar articles
- The basic helix-loop-helix transcription factor dHAND, a marker gene for the developing human sympathetic nervous system, is expressed in both high- and low-stage neuroblastomas.
Gestblom C, Grynfeld A, Ora I, Ortoft E, Larsson C, Axelson H, Sandstedt B, Cserjesi P, Olson EN, Påhlman S. Gestblom C, et al. Lab Invest. 1999 Jan;79(1):67-79. Lab Invest. 1999. PMID: 9952112 - Meta-mining of neuroblastoma and neuroblast gene expression profiles reveals candidate therapeutic compounds.
De Preter K, De Brouwer S, Van Maerken T, Pattyn F, Schramm A, Eggert A, Vandesompele J, Speleman F. De Preter K, et al. Clin Cancer Res. 2009 Jun 1;15(11):3690-6. doi: 10.1158/1078-0432.CCR-08-2699. Epub 2009 May 12. Clin Cancer Res. 2009. PMID: 19435837 - Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma.
Jansky S, Sharma AK, Körber V, Quintero A, Toprak UH, Wecht EM, Gartlgruber M, Greco A, Chomsky E, Grünewald TGP, Henrich KO, Tanay A, Herrmann C, Höfer T, Westermann F. Jansky S, et al. Nat Genet. 2021 May;53(5):683-693. doi: 10.1038/s41588-021-00806-1. Epub 2021 Mar 25. Nat Genet. 2021. PMID: 33767450 - Role of PACAP in the physiology and pathology of the sympathoadrenal system.
Ghzili H, Grumolato L, Thouënnon E, Tanguy Y, Turquier V, Vaudry H, Anouar Y. Ghzili H, et al. Front Neuroendocrinol. 2008 Jan;29(1):128-41. doi: 10.1016/j.yfrne.2007.10.001. Epub 2007 Oct 22. Front Neuroendocrinol. 2008. PMID: 18048093 Review. - [New insights into the genetic basis of neuroblastoma].
Speleman F, De Preter K, Hoebeeck J, Van Roy N, Vandesompele J. Speleman F, et al. Verh K Acad Geneeskd Belg. 2007;69(4):167-96. Verh K Acad Geneeskd Belg. 2007. PMID: 17821957 Review. Dutch.
Cited by
- Meta-analysis of muscle transcriptome data using the MADMuscle database reveals biologically relevant gene patterns.
Baron D, Dubois E, Bihouée A, Teusan R, Steenman M, Jourdon P, Magot A, Péréon Y, Veitia R, Savagner F, Ramstein G, Houlgatte R. Baron D, et al. BMC Genomics. 2011 Feb 16;12:113. doi: 10.1186/1471-2164-12-113. BMC Genomics. 2011. PMID: 21324190 Free PMC article. - Involvement of GTA protein NC2beta in neuroblastoma pathogenesis suggests that it physiologically participates in the regulation of cell proliferation.
Di Pietro C, Ragusa M, Barbagallo D, Duro LR, Guglielmino MR, Majorana A, Giunta V, Rapisarda A, Tricarichi E, Miceli M, Angelica R, Grillo A, Banelli B, Defferari I, Forte S, Laganà A, Bosco C, Giugno R, Pulvirenti A, Ferro A, Grzeschik KH, Di Cataldo A, Tonini GP, Romani M, Purrello M. Di Pietro C, et al. Mol Cancer. 2008 Jun 6;7:52. doi: 10.1186/1476-4598-7-52. Mol Cancer. 2008. PMID: 18538002 Free PMC article. - Promoter hypermethylation as a mechanism for Lamin A/C silencing in a subset of neuroblastoma cells.
Rauschert I, Aldunate F, Preussner J, Arocena-Sutz M, Peraza V, Looso M, Benech JC, Agrelo R. Rauschert I, et al. PLoS One. 2017 Apr 19;12(4):e0175953. doi: 10.1371/journal.pone.0175953. eCollection 2017. PLoS One. 2017. PMID: 28422997 Free PMC article. - IgA antibody immunotherapy targeting GD2 is effective in preclinical neuroblastoma models.
Stip MC, Evers M, Nederend M, Chan C, Reiding KR, Damen MJ, Heck AJR, Koustoulidou S, Ramakers R, Krijger GC, de Roos R, Souteyrand E, Cornel AM, Dierselhuis MP, Jansen M, de Boer M, Valerius T, van Tetering G, Leusen JHW, Meyer-Wentrup F. Stip MC, et al. J Immunother Cancer. 2023 Jul;11(7):e006948. doi: 10.1136/jitc-2023-006948. J Immunother Cancer. 2023. PMID: 37479484 Free PMC article.
References
- Berwanger B, Hartmann O, Bergmann E, Bernard S, Nielsen D, Krause M, Kartal A, Flynn D, Wiedemeyer R, Schwab M, et al. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell. 2002;2(5):377–386. doi: 10.1016/S1535-6108(02)00179-4. - DOI - PubMed
- McArdle L, McDermott M, Purcell R, Grehan D, O'Meara A, Breatnach F, Catchpoole D, Culhane AC, Jeffery I, Gallagher WM, et al. Oligonucleotide microarray analysis of gene expression in neuroblastoma displaying loss of chromosome 11q. Carcinogenesis. 2004 - PubMed
- Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, Seiser E, Jagannathan J, Shusterman S, Bansal M, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer research. 2006;66(12):6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical