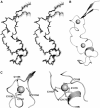Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase - PubMed (original) (raw)
Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase
Mark D Allen et al. EMBO J. 2006.
Abstract
Methylation of CpG dinucleotides is the major epigenetic modification of mammalian genomes, critical for regulating chromatin structure and gene activity. The mixed-lineage leukaemia (MLL) CXXC domain selectively binds nonmethyl-CpG DNA, and is required for transformation by MLL fusion proteins that commonly arise from recurrent chromosomal translocations in infant and secondary treatment-related acute leukaemias. To elucidate the molecular basis of nonmethyl-CpG DNA recognition, we determined the structure of the human MLL CXXC domain by multidimensional NMR spectroscopy. The CXXC domain has a novel fold in which two zinc ions are each coordinated tetrahedrally by four conserved cysteine ligands provided by two CGXCXXC motifs and two distal cysteine residues. We have identified the CXXC domain DNA binding interface by means of chemical shift perturbation analysis, cross-saturation transfer and site-directed mutagenesis. In particular, we have shown that residues in an extended surface loop are in close contact with the DNA. These data provide a template for the design of specifically targeted therapeutics for poor prognosis MLL-associated leukaemias.
Figures
Figure 1
Solution structure of the MLL CXXC domain. (A) An overlay of the backbone atoms of the 20 lowest energy structures in stereo. (B) A ribbon representation of the lowest energy structure (same orientation as in (A)), prepared using the program PyMOL (
). Zn ions are shown as spheres. (C) A ribbon representation of the Zn coordination sites in MLL (PyMOL).
Figure 2
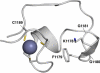
Ribbon representation of the elaborate turn in the CXXC domain of MLL showing the side chains of the residues from the KFGG motif and the second Zn coordination site (PyMOL). An extended loop is formed between residues G1181 and C1189.
Figure 3
Structure based sequence alignment of representative CXXC domains. Sequences were aligned using Jalview (Clamp et al, 2004) and are shaded in blue according to the degree of amino-acid sequence identity. All sequences are from Homo sapiens except where indicated. The secondary structure elements are labelled α for helix and β for strands, as calculated by DSSP (Kabsch and Sander, 1983). Filled circle (•) and filled diamonds (⧫) indicate cysteines involved in the first and second Zn coordination sites, respectively. NCBI accession numbers are as follows: MLL gi:56550039; MLL (F. rubripes) gi:3309542; MLL4 gi:7662046; DNMT1 gi:4503351; FLX10 gi:54112382; CXCC1/CGBP gi:7656975; MBD1 gi:21464117; LCX gi:33859755.
Figure 4
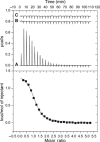
ITC analysis of DNA binding by the MLL CXXC domain. Typical ITC data are shown for the endothermic binding of the CXXC domain to a 12-mer CpG-containing DNA oligonucleotide at 22°C in 20 mM MES, pH 6.5, 250 mM NaCl, 5 mM β-mercaptoethanol. Upper panel: (A) CXXC domain (1.3 mM) into the calorimetric cell (1.4 ml) containing CpG 12-mer DNA (49 μM). (B) CXXC domain (970 μM) into ITC buffer. (C) ITC buffer into CpG 12-mer DNA (49 μM). Lower Panel: Integrated heat pulses, normalised per mole of injectant, giving a differential binding curve that is adequately described by a one-site binding model.
Figure 5
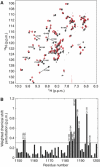
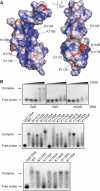
Chemical shift perturbation analysis of DNA binding by the MLL CXXC domain. (A) An overlay of the 1H–15N HSQC spectra of the CXXC domain in the absence (black contour levels) or the presence (red contour levels) of an equimolar concentration of the palindromic 12-mer oligonucleotide GTATCCGGATAC. This is a combined 15N 1H chemical shift perturbation map as defined by Δ1H+(Δ15N/5) (Hajduk et al, 1997). Chemical shift perturbations greater than 0.3 p.p.m. are indicated as lines connecting the amide resonances in the free and bound states. (B) A plot of chemical shift due to DNA binding for residues R1150–P1201, with a cutoff at 0.3 p.p.m.
Figure 6
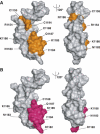
Mapping the DNA binding surface of the MLL CXXC domain. (A) Representations of the molecular surface of the MLL CXXC domain, with two views related by a rotation of 120° about the vertical axis. Residues that undergo significant (>0.3 p.p.m.) chemical shift upon binding DNA are coloured orange. (B) Representations of the molecular surface of the MLL CXXC domain. The two views are related by a rotation of 120° about the vertical axis. Residues that show a decrease in peak intensity of >15% upon saturation of the imino protons of the DNA and a mixing time of 1.44 s are coloured pink.
Figure 7
Mutational analysis of DNA binding by the MLL CXXC domain. (A) Representation of the electrostatic surface potential of the CXXC domain as calculated by the program APBS (Baker et al, 2001) and coloured using a linear colour ramp from −25.0 kT (red) to +25.0 kT (blue). Residues that are functionally implicated in DNA binding in gel shift assays are indicated. (B) Gel shift assays. Purified 6xHis-tagged proteins were incubated with 12-mer dsDNA carrying a methyl- or nonmethyl-CpG pair and electrophoresed in agarose gel shift assays as shown. The wild type and mutant proteins utilised are indicated.
Similar articles
- Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia.
Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Cierpicki T, et al. Nat Struct Mol Biol. 2010 Jan;17(1):62-8. doi: 10.1038/nsmb.1714. Epub 2009 Dec 13. Nat Struct Mol Biol. 2010. PMID: 20010842 Free PMC article. - Functional specificity of CpG DNA-binding CXXC domains in mixed lineage leukemia.
Risner LE, Kuntimaddi A, Lokken AA, Achille NJ, Birch NW, Schoenfelt K, Bushweller JH, Zeleznik-Le NJ. Risner LE, et al. J Biol Chem. 2013 Oct 11;288(41):29901-10. doi: 10.1074/jbc.M113.474858. Epub 2013 Aug 29. J Biol Chem. 2013. PMID: 23990460 Free PMC article. - Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2.
Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK. Bach C, et al. Oncogene. 2009 Feb 12;28(6):815-23. doi: 10.1038/onc.2008.443. Epub 2008 Dec 8. Oncogene. 2009. PMID: 19060922 - MLL translocations, histone modifications and leukaemia stem-cell development.
Krivtsov AV, Armstrong SA. Krivtsov AV, et al. Nat Rev Cancer. 2007 Nov;7(11):823-33. doi: 10.1038/nrc2253. Nat Rev Cancer. 2007. PMID: 17957188 Review. - The molecular biology of mixed lineage leukemia.
Slany RK. Slany RK. Haematologica. 2009 Jul;94(7):984-93. doi: 10.3324/haematol.2008.002436. Epub 2009 Jun 16. Haematologica. 2009. PMID: 19535349 Free PMC article. Review.
Cited by
- H3K4me2 distinguishes a distinct class of enhancers during the maternal-to-zygotic transition.
Hurton MD, Miller JM, Lee MT. Hurton MD, et al. bioRxiv [Preprint]. 2024 Aug 26:2024.08.26.609713. doi: 10.1101/2024.08.26.609713. bioRxiv. 2024. PMID: 39253505 Free PMC article. Preprint. - Menin as a hub controlling mixed lineage leukemia.
Thiel AT, Huang J, Lei M, Hua X. Thiel AT, et al. Bioessays. 2012 Sep;34(9):771-80. doi: 10.1002/bies.201200007. Epub 2012 Jul 24. Bioessays. 2012. PMID: 22829075 Free PMC article. Review. - Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia.
Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Cierpicki T, et al. Nat Struct Mol Biol. 2010 Jan;17(1):62-8. doi: 10.1038/nsmb.1714. Epub 2009 Dec 13. Nat Struct Mol Biol. 2010. PMID: 20010842 Free PMC article. - The pathogenesis of mixed-lineage leukemia.
Muntean AG, Hess JL. Muntean AG, et al. Annu Rev Pathol. 2012;7:283-301. doi: 10.1146/annurev-pathol-011811-132434. Epub 2011 Oct 17. Annu Rev Pathol. 2012. PMID: 22017583 Free PMC article. Review. - Structure, Activity and Function of the MLL2 (KMT2B) Protein Lysine Methyltransferase.
Klonou A, Chlamydas S, Piperi C. Klonou A, et al. Life (Basel). 2021 Aug 12;11(8):823. doi: 10.3390/life11080823. Life (Basel). 2021. PMID: 34440566 Free PMC article. Review.
References
- Bax A (1994) Multidimensional nuclear-magnetic-resonance methods for protein studies. Curr Opin Struct Biol 4: 738–744
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases