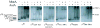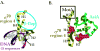Mutational analysis of sigma70 region 4 needed for appropriation by the bacteriophage T4 transcription factors AsiA and MotA - PubMed (original) (raw)
Mutational analysis of sigma70 region 4 needed for appropriation by the bacteriophage T4 transcription factors AsiA and MotA
Kimberly Baxter et al. J Mol Biol. 2006.
Abstract
Transcriptional activation of bacteriophage T4 middle promoters requires sigma70-containing Escherichia coli RNA polymerase, the T4 activator MotA, and the T4 co-activator AsiA. T4 middle promoters contain the sigma70 -10 DNA element. However, these promoters lack the sigma70 -35 element, having instead a MotA box centered at -30, which is bound by MotA. Previous work has indicated that AsiA and MotA interact with region 4 of sigma70, the C-terminal portion that normally contacts -35 DNA and the beta-flap structure in core. AsiA binding prevents the sigma70/beta-flap and sigma70/-35 DNA interactions, inhibiting transcription from promoters that require a -35 element. To test the importance of residues within sigma70 region 4 for MotA and AsiA function, we investigated how sigma70 region 4 mutants interact with AsiA, MotA, and the beta-flap and function in transcription assays in vitro. We find that alanine substitutions at residues 584-588 (region 4.2) do not impair the interaction of region 4 with the beta-flap or MotA, but they eliminate the interaction with AsiA and prevent AsiA inhibition and MotA/AsiA activation. In contrast, alanine substitutions at 551-552, 554-555 (region 4.1) eliminate the region 4/beta-flap interaction, significantly impair the AsiA/sigma70 interaction, and eliminate AsiA inhibition. However, the 4.1 mutant sigma70 is still fully competent for activation if both MotA and AsiA are present. A previous NMR structure shows AsiA binding to sigma70 region 4, dramatically distorting regions 4.1 and 4.2 and indirectly changing the conformation of the MotA interaction site at the sigma70 C terminus. Our analyses provide biochemical relevance for the sigma70 residues identified in the structure, indicate that the interaction of AsiA with sigma70 region 4.2 is crucial for activation, and support the idea that AsiA binding facilitates an interaction between MotA and the far C terminus of sigma70.
Figures
Fig. 1
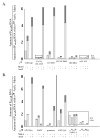
Region 4 of wild type σ70 and mutant σ proteins and the T4 middle promoter PuvsX. A. The C-terminal amino acid sequence of σ70 is shown from residues 540 to 613. Residues in pink contact the -35 element of E. coli promoter DNA while residues in blue contact the β-flap tip ; ; ; . Every tenth residue is marked with a small vertical line. Above the sequence is shown the locations of regions 4.1 and 4.2 and the positions of the α-helices H1, H2, and H5 and the helix-turn-helix H3-T-H4 structures observed in region 4 of the primary σ of T. aquaticus complexed with -35 element DNA . Residues that contact AsiA in the AsiA/σ70 region 4 structure are designated with an asterisk and the region of the MotA contact site is indicated. Below are shown the region 4 sequences of the mutant σ proteins, with residues that differ from σ70 in red. B. The sequence of PuvsX from −61 to +9 is shown with the start of transcription indicated as +1. The locations of the σ70 –10 element and the MotA box are indicated. Sequences within PuvsX that match the canonical polymerase elements σ70 -10 element (TATAAT); σ70 extended -10 element (-15T, -14G); σ70 -35 element (TTGACA); and α UP element (AAAa/ta/tTa/tTTTTnnAAAA) are in red.
Fig. 2
Transcriptional activities of polymerases reconstituted with mutant σ proteins using PuvsX in vitro in the presence of AsiA and/or MotA. The indicated σ (0.2 pmol) and AsiAHis6 (3 pmol ) were incubated in 1.8 μl of protein buffer I for 10 min at 37° C. A solution containing 0.05 pmol of core in 1.1 μl of protein buffer II was then added, and the resulting solution was incubated for an additional 10 min at 37° C. Transcription was initiated by adding this solution to 2.1 μl of DNA buffer containing 0.01 pmol DNA. After 37° C for 20 sec, rifampicin (150 ng) was added to limit transcription to a single round. For each indicated σ, [the amount of PuvsX RNA observed]/[the amount of PusvX RNA observed with σ70 polymerase in the absence of MotA and AsiA] is shown using polymerase alone, polymerase with MotA, polymerase with AsiA, and polymerase with AsiA and MotA. Values were determined from two or more transcriptions. Error is indicated by darker shaded area. In some cases where the level of transcription was low, 10-fold values (10 × values) are also shown. In these cases, zero is the bottom of the enclosed box.
Fig. 3
σ/AsiA, σ/MotA, and AsiA/σ/MotA complexes in native protein gels. The indicated σ was incubated with excess His6AsiA and/or MotA and then subjected to electrophoresis through native 6 % acrylamide gels as described in Materials and Methods. The positions of σ, the σ/AsiA complex, the σ/MotA complex, and the AsiA/σ/MotA complex are shown. The positions of free MotA and AsiA are also marked. Excess free MotA protein is the dark band that migrates near the top of the gel in the lanes with added MotA. The excess free AsiA is usually not seen because AsiA is a small protein (10 kDa) and stains poorly.
Fig. 4
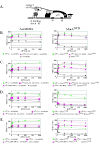
2-hybrid assay and the effect of σ70 substitutions on the interaction of σ70 region 4 with the AsiAK20A and MotANTD. (A) Cartoon shows the test promoter, which has a λ operator (OR) located 62 bp upstream from the initiation site of the lac core promoter. The presence of the α-σ chimera protein, expressed by a pBRα-σ plasmid, and the cI-bait fusion protein, expressed by a pcI-bait plasmid, can result in an increase in_lacZ_ transcription and subsequent β-galactosidase activity if there is an interaction between the σ present in the chimera and the bait protein. The addition of IPTG induces expression of both the α-σ chimera and the cI-bait fusion protein. B-E. Effect of σ70 mutations on the interaction of σ70 region 4 with AsiAK20A (left panels) and MotANTD (right panels). Cultures contained the pcI-AsiAK20A or pcI-MotANTD plasmid and either the control plasmid, pBRα that lacks σ70 region 4, or as indicated, a pBRα-σ70 plasmid that has σ70 region 4 (σ70 4), σ70 region 4.1 (σ70 4.1), σ70 region 4.2 (σ70 4.2) with or without the indicated σ70 substitutions. Graphs show β-galactosidase activity (in Miller units) versus the concentration of IPTG present in the cultures.
Fig. 5
The σ70 substitutions 551-552,554-555A (A) eliminate the interaction of the β-flap with σ70 region 4 whereas substitutions 584-588A (B) do not impair the interaction of σ70 region 4 with the β-flap. Cultures contained a pBRα-σ70 plasmid, which had σ70 region 4 (σ70 4) with the indicated substitutions, and either the control plasmid, pACλcI32, the pcI plasmid that lacks a fused bait protein, or the pcI-β-flap plasmid (β-flap). Graphs show β-galactosidase activity (in Miller units) versus the concentration of IPTG present in the cultures.
Fig. 6
Interaction of σ70 region 4 with -35 element and the β-flap versus its interaction with AsiA and MotA. Structures of (A) T. aquaticus σ region 4 (residues corresponding to E. coli σ70 residues 546-612) with the –35 region of promoter DNA (PDB: 1KU7) and of (B) E. coli σ70 region 4 (residues 546-613) with AsiA (residues 2-90) (PDB: 1TLH). σ70 region 4 is shown in gold, AsiA is shown in green, and the DNA is shown in magenta. Alpha helices H2, H3, H4, and H5 in σ70 (see Fig. 1A) are labeled. Spacefill structures of σ70 residues 551, 554, and 555 in region 4.1 and residues 584 and 587 in region 4.2, which are predicted to interact with AsiA in the structure and whose substitution impairs the σ70/AsiA interaction, are shown in red in (B). The portion of σ70 region 4 that interacts with the β-flap is indicated by the blue oval in (A). H5 of σ70 that interacts with MotA is indicated by the black rectangle in (B). Note that the binding of AsiA to σ70 region 4 converts H3-T-H4 of σ70, which normally interacts with the –35 sequences of promoter DNA, into one continuous helix.
Similar articles
- Transcriptional control in the prereplicative phase of T4 development.
Hinton DM. Hinton DM. Virol J. 2010 Oct 28;7:289. doi: 10.1186/1743-422X-7-289. Virol J. 2010. PMID: 21029433 Free PMC article. Review. - A family of anti-sigma70 proteins in T4-type phages and bacteria that are similar to AsiA, a Transcription inhibitor and co-activator of bacteriophage T4.
Pineda M, Gregory BD, Szczypinski B, Baxter KR, Hochschild A, Miller ES, Hinton DM. Pineda M, et al. J Mol Biol. 2004 Dec 10;344(5):1183-97. doi: 10.1016/j.jmb.2004.10.003. J Mol Biol. 2004. PMID: 15561138 - Bacteriophage T4 MotA activator and the β-flap tip of RNA polymerase target the same set of σ70 carboxyl-terminal residues.
Bonocora RP, Decker PK, Glass S, Knipling L, Hinton DM. Bonocora RP, et al. J Biol Chem. 2011 Nov 11;286(45):39290-6. doi: 10.1074/jbc.M111.278762. Epub 2011 Sep 12. J Biol Chem. 2011. PMID: 21911499 Free PMC article. - Transcriptional takeover by sigma appropriation: remodelling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and co-activator AsiA.
Hinton DM, Pande S, Wais N, Johnson XB, Vuthoori M, Makela A, Hook-Barnard I. Hinton DM, et al. Microbiology (Reading). 2005 Jun;151(Pt 6):1729-1740. doi: 10.1099/mic.0.27972-0. Microbiology (Reading). 2005. PMID: 15941982 Review.
Cited by
- The Xp10 Bacteriophage Protein P7 Inhibits Transcription by the Major and Major Variant Forms of the Host RNA Polymerase via a Common Mechanism.
Brown DR, Sheppard CM, Burchell L, Matthews S, Wigneshweraraj S. Brown DR, et al. J Mol Biol. 2016 Oct 9;428(20):3911-3919. doi: 10.1016/j.jmb.2016.08.004. Epub 2016 Aug 8. J Mol Biol. 2016. PMID: 27515396 Free PMC article. - Activation of transcription initiation by Spx: formation of transcription complex and identification of a Cis-acting element required for transcriptional activation.
Reyes DY, Zuber P. Reyes DY, et al. Mol Microbiol. 2008 Aug;69(3):765-79. doi: 10.1111/j.1365-2958.2008.06330.x. Mol Microbiol. 2008. PMID: 18687074 Free PMC article. - Transcriptional control in the prereplicative phase of T4 development.
Hinton DM. Hinton DM. Virol J. 2010 Oct 28;7:289. doi: 10.1186/1743-422X-7-289. Virol J. 2010. PMID: 21029433 Free PMC article. Review. - A single rare σ70 variant establishes a unique gene expression pattern in the E. coli pathobiont LF82.
Arroyo-Mendoza M, Proctor A, Correa-Medina A, DeWolf S, Brand MW, Rosas V, Lorenzi H, Wannemuehler MJ, Phillips GJ, Hinton DM. Arroyo-Mendoza M, et al. Nucleic Acids Res. 2024 Oct 28;52(19):11552-11570. doi: 10.1093/nar/gkae773. Nucleic Acids Res. 2024. PMID: 39258538 Free PMC article. - Direct activator/co-activator interaction is essential for bacteriophage T4 middle gene expression.
Yuan AH, Hochschild A. Yuan AH, et al. Mol Microbiol. 2009 Nov;74(4):1018-30. doi: 10.1111/j.1365-2958.2009.06916.x. Epub 2009 Oct 15. Mol Microbiol. 2009. PMID: 19843221 Free PMC article.
References
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–66. - PubMed
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources