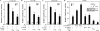LZTS2 is a novel beta-catenin-interacting protein and regulates the nuclear export of beta-catenin - PubMed (original) (raw)
LZTS2 is a novel beta-catenin-interacting protein and regulates the nuclear export of beta-catenin
Gregory Thyssen et al. Mol Cell Biol. 2006 Dec.
Abstract
Beta-catenin plays multiple roles in cell-cell adhesion and Wnt signal transduction. Through the Wnt signal, the cellular level of beta-catenin is constitutively regulated by the multicomponent destruction complex containing glycogen synthase kinase 3beta, axin, and adenomatous polyposis coli. Here, we present multiple lines of evidence to demonstrate that LZTS2 (lucine zipper tumor suppressor 2) interacts with beta-catenin, represses the transactivation of beta-catenin, and affects the subcellular localization of beta-catenin. The LZTS2 gene is located at 10q24.3, which is frequently lost in a variety of human tumors. A functional nuclear export signal (NES) was identified in the C terminus of the protein (amino acids 631 to 641). Appending this motif to green fluorescent protein (GFP) induced nuclear exclusion of the GFP fusion protein. However, introducing point mutations in either one or two leucine residues of this NES sequence abolished the nuclear exclusion of the LZTS2 protein. The nuclear export of LZTS2 can be blocked by leptomycin B (LMB), an inhibitor of the CRM1/exportin-alpha pathway. Intriguingly, beta-catenin colocalizes with LZTS2 in the cytoplasm of cells in the absence of LMB but in the nuclei of cells in the presence of LMB. Increasing the LZTS2 protein in cells reduces the level of nuclear beta-catenin in SW480 cells. Taken together, these data demonstrate that LZTS2 is a beta-catenin-interacting protein that can modulate beta-catenin signaling and localization.
Figures
FIG. 1.
Alignment of LZTS proteins. (A) Alignment of the three LZTS protein sequences, including LZTS1, also called FEZ1 (AAD23840), LZTS2, also called LAPSER1/KIAA1813 (AAH58938), and LZTS3, also called ProSapip1 (AAH38860). Identical and similar residues are highlighted in black and gray, respectively. Asterisks denote leucine or similar hydrophobic residues with spacing consistent with leucine zipper domains. The NES sequence characterized in the text is underlined. (B) Alignment of similar NES sites in orthologs of hLZTS2 (AAH58938) from Xenopus tropicalis (AAH75457), Mus musculus (NP_663478), Canis familiaris (XP_543975), and Pan troglodytes (XP_521656).
FIG. 2.
LZTS2 interacts with β-catenin in cells. (A) A schematic representation of the yeast two-hybrid assay for mapping the interaction between the β-catenin and LZTS2 proteins. (B) The cDNA fragments containing different portions of human LZTS2 were fused to a GAL4 transactivation domain, and the armadillo repeats of β-catenin were fused to the GAL4 DNA binding domain in pGBT9 vector. Numbers correspond to amino acid residues. Both of the plasmids were cotransformed into PJ69-4A cells as labeled in the figure and plated on SD-Ade-Leu-Trp plates or SD-Leu-Trp plates to monitor transformation efficiency. Three independent colonies were inoculated from each transformation for subsequent liquid β-Gal assays. The data for the liquid β-Gal assays are reported in relative units normalized by cell number (optical density at 600 nm [OD 600]). (C) The indicated fragments of β-catenin were examined by yeast two-hybrid assay for interaction with the C-terminal fragment of hLZTS2 (aa 447 to 669). (D) Equal amounts of GST-β-catenin fusion proteins were immobilized on a glutathione-Sepharose matrix. The binding of [35S]methionine-labeled LZTS2 to GST-β-catenin fusion proteins was analyzed by SDS-PAGE and visualized by autoradiography. (E and F) Whole-cell lysates of CV-1 cells transfected with full-length β-catenin and FLAG-tagged LZTS2 were immunoprecipitated with normal mouse IgG or a β-catenin (E) or FLAG (F) monoclonal antibody. The immunoprecipitates were analyzed by Western blotting with different antibodies as indicated. (G) Whole-cell lysates of SW480 cells were immunoprecipitated with the homemade LZTS2 antibody and normal rabbit IgG and analyzed by Western blotting.
FIG. 3.
LZTS2 represses β-catenin-mediated transcription. (A) One hundred nanograms of pGL3-OT (OT) or pGL3-OF (OF), 25 ng of pcDNA3-β-gal, 5 ng of TCF1 expression vector, 20 ng of β-catenin, and various amounts of pcDNA3-FLAG-hLZTS2 as indicated were transfected into LNCaP cells. Cells were cultured for 24 h in the regular media and luciferase, and β-Gal activities were measured as indicated above. Similar experiments were repeated with PC3 cells (B) and DU145 cells (C). (D) Either 100 ng of pGL3-OT (OT) or pGL3-OF (OF), 25 ng of pcDNA3-β-gal, and other shRNA or expression constructs as indicated in the figure were transfected into the human colon cancer cell line SW480. Luciferase and β-Gal activities were measured in panel C.
FIG. 4.
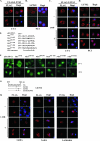
LZTS2 localizes in the cytoplasm and contains a leptomycin B-regulated NES motif. (A) The pcDNA3-FLAG-hLZTS2 expression vector was transfected into CV-1 cells. The ectopically expressed hLZTS2 was detected with FLAG monoclonal antibody and revealed by rhodamine-conjugated secondary antibody (red). The nuclei were counterstained with DAPI (blue). (B) Endogenous hLZTS2 was detected with a rabbit antibody in PC3 cells. (C) CV-1 cells were transfected with the pcDNA3-FLAG-hLZTS2 expression vector and then incubated in medium either with or without 60 ng/ml LMB. The ectopically expressed hLZTS2 was detected as in panel A. (D) PC3 cells were cultured in medium either with or without 60 ng/ml LMB for 24 h and then stained with the antibody against endogenous hLZTS2 (red). (E) The GFP fusion proteins containing the putative NES sequences of hLZTS2 and an HIV-Rev classical NES were generated in the pEGFP-C1 vector. Numbers correspond to amino acid residues. (F) The expression vectors containing the above GFP fusion proteins were transfected into CV-1 and PC3 cells. After 24 h of transfection, live cells were observed with an inverted fluorescence microscope. (G) A schematic representation of the NES sequence in hLZTS2. Either single or double mutations within the site were introduced. (H) The expression vectors of pcDNA3-FLAG-hLZTS2 containing either single or double point mutations were transfected into CV-1 cells. The cells were treated with LMB as indicated in panel C. The localization of the LZTS2 proteins was detected as in panel A.
FIG. 5.
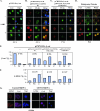
LZTS2 affects the cellular localization of β-catenin. (A) pcDNA3-HA-β-catenin was transfected into CV-1 cells and incubated with or without 60 ng/ml LMB. The subcellular localization of the β-catenin (β-cat) was monitored by hemagglutinin monoclonal antibody and fluorescein isothiocyanate-conjugated secondary antibody (green). (B) Both pcDNA3-HA-β-catenin and pcDNA3-FLAG-hLZTS2 were transfected into CV-1 cells. Cells were then cultured with or without 60 ng/ml LMB for 24 h. The ectopically expressed proteins were detected with FLAG or hemagglutinin antibody and revealed with rhodamine or fluorescein isothiocyanate-conjugated secondary antibody, respectively. (C) As described above, the mutant of LZTS2 was used in the experiment to examine the colocalization with β-catenin. (D) PC3 cells were cultured in medium either with or without 60 ng/ml LMB. Endogenous β-catenin and LZTS2 proteins were detected by the specific antibodies against each protein and revealed with appropriate secondary antibodies. (E and F) Percentages of cells in which the localization of the β-catenin and LZST2 proteins in the nuclei (N), cytoplasm (C), or both (N and C) were assessed in CV-1 cells as described in the above experiments. (G) SW480 cells were transfected with either the pBS/U6-LZTS shRNA or pBS/U6 vector as a negative control and then fixed and stained for endogenous LZTS2 and β-catenin after 48 h. The rabbit anti-LZTS2 or mouse anti-β-catenin antibody was developed with the Alexa-Fluor goat anti-rabbit 594 (Invitrogen) (red) or Alexa-Fluor donkey anti-mouse 647 (pink) antibody, respectively. DAPI was used for nuclear visualization.
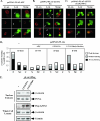
FIG. 6.
LZTS2 regulates the cellular level of β-catenin. The FLAG-tagged expression vectors containing wild-type APC (A) or LZTS2 (B) or the LZTS2 NES mutant (C) were transfected into SW480 cells. Cells were stained with FLAG and β-catenin antibodies for the ectopically expressed proteins and endogenous β-catenin, as indicated by arrows. (D) Percentages of positively stained β-catenin cells were measured from the above experiments. The intensity and localization of β-catenin staining in the nuclei (N), cytoplasm (C), or both (N and C) were analyzed in cells transfected with different expression constructs as indicated. (E) SW480 cells were infected with the pLentiviral vectors for either wild-type or mutated LZTS2. The nuclear extracts and whole-cell lysates were prepared from the above cells and used for Western blotting.
Similar articles
- LZTS2 and PTEN collaboratively regulate ß-catenin in prostatic tumorigenesis.
Yu EJ, Hooker E, Johnson DT, Kwak MK, Zou K, Luong R, He Y, Sun Z. Yu EJ, et al. PLoS One. 2017 Mar 21;12(3):e0174357. doi: 10.1371/journal.pone.0174357. eCollection 2017. PLoS One. 2017. PMID: 28323888 Free PMC article. - Lzts2 regulates embryonic cell movements and dorsoventral patterning through interaction with and export of nuclear β-catenin in zebrafish.
Li Y, Li Q, Long Y, Cui Z. Li Y, et al. J Biol Chem. 2011 Dec 30;286(52):45116-30. doi: 10.1074/jbc.M111.267328. Epub 2011 Nov 4. J Biol Chem. 2011. PMID: 22057270 Free PMC article. - The leucine zipper putative tumor suppressor 2 protein LZTS2 regulates kidney development.
Peng Y, Clark C, Luong R, Tu WH, Lee J, Johnson DT, Das A, Carroll TJ, Sun Z. Peng Y, et al. J Biol Chem. 2011 Nov 18;286(46):40331-42. doi: 10.1074/jbc.M111.302059. Epub 2011 Sep 26. J Biol Chem. 2011. PMID: 21949185 Free PMC article. - Calcium, calcium-sensing receptor and colon cancer.
Whitfield JF. Whitfield JF. Cancer Lett. 2009 Mar 8;275(1):9-16. doi: 10.1016/j.canlet.2008.07.001. Epub 2008 Aug 23. Cancer Lett. 2009. PMID: 18725175 Review. - Looking beyond the Wnt pathway for the deep nature of β-catenin.
Fagotto F. Fagotto F. EMBO Rep. 2013 May;14(5):422-33. doi: 10.1038/embor.2013.45. Epub 2013 Apr 19. EMBO Rep. 2013. PMID: 23598517 Free PMC article. Review.
Cited by
- Long non-coding RNA linc00921 suppresses tumorigenesis and epithelial-to-mesenchymal transition of triple-negative breast cancer via targeting miR-9-5p/LZTS2 axis.
Zhang J, Zhang L, Wang J, Zhao J, Zhao X, Zhang C, Han P, Geng C. Zhang J, et al. Hum Cell. 2022 May;35(3):909-923. doi: 10.1007/s13577-022-00685-6. Epub 2022 Feb 18. Hum Cell. 2022. PMID: 35179718 Free PMC article. - Cancer stem cells: a major culprit of intra-tumor heterogeneity.
Naz F, Shi M, Sajid S, Yang Z, Yu C. Naz F, et al. Am J Cancer Res. 2021 Dec 15;11(12):5782-5811. eCollection 2021. Am J Cancer Res. 2021. PMID: 35018226 Free PMC article. Review. - Physiological role of β-catenin/TCF signaling in neurons of the adult brain.
Wisniewska MB. Wisniewska MB. Neurochem Res. 2013 Jun;38(6):1144-55. doi: 10.1007/s11064-013-0980-9. Epub 2013 Feb 2. Neurochem Res. 2013. PMID: 23377854 Free PMC article. Review. - Nucleoporin 62-like protein activates canonical Wnt signaling through facilitating the nuclear import of β-catenin in zebrafish.
Yang X, Gu Q, Lin L, Li S, Zhong S, Li Q, Cui Z. Yang X, et al. Mol Cell Biol. 2015 Apr;35(7):1110-24. doi: 10.1128/MCB.01181-14. Epub 2015 Jan 20. Mol Cell Biol. 2015. PMID: 25605329 Free PMC article. - Interactome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor.
Pilot-Storck F, Chopin E, Rual JF, Baudot A, Dobrokhotov P, Robinson-Rechavi M, Brun C, Cusick ME, Hill DE, Schaeffer L, Vidal M, Goillot E. Pilot-Storck F, et al. Mol Cell Proteomics. 2010 Jul;9(7):1578-93. doi: 10.1074/mcp.M900568-MCP200. Epub 2010 Apr 5. Mol Cell Proteomics. 2010. PMID: 20368287 Free PMC article.
References
- Cabeza-Arvelaiz, Y., T. C. Thompson, J. L. Sepulveda, and A. C. Chinault. 2001. LAPSER1: a novel candidate tumor suppressor gene from 10q24.3. Oncogene 20:6707-6717. - PubMed
- Chan, S. K., and G. Struhl. 2002. Evidence that Armadillo transduces wingless by mediating nuclear export or cytosolic activation of Pangolin. Cell 111:265-280. - PubMed
- Daniels, D. L., and W. I. Weis. 2002. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol. Cell 10:573-584. - PubMed
- Dignam, J. D., P. L. Martin, B. S. Shastry, and R. G. Roeder. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582-598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA 87767/CA/NCI NIH HHS/United States
- R01 DK061002-03/DK/NIDDK NIH HHS/United States
- R01 CA070297/CA/NCI NIH HHS/United States
- R01 CA087767-02/CA/NCI NIH HHS/United States
- DK 61002/DK/NIDDK NIH HHS/United States
- R01 DK061002-01A1/DK/NIDDK NIH HHS/United States
- R01 CA087767-04/CA/NCI NIH HHS/United States
- R01 CA070297-08/CA/NCI NIH HHS/United States
- R01 CA070297-07/CA/NCI NIH HHS/United States
- R01 CA070297-11A2/CA/NCI NIH HHS/United States
- R01 CA087767-03/CA/NCI NIH HHS/United States
- Z01 DK061002/ImNIH/Intramural NIH HHS/United States
- R01 DK061002-04/DK/NIDDK NIH HHS/United States
- R56 DK061002-06A1/DK/NIDDK NIH HHS/United States
- R01 CA087767-05/CA/NCI NIH HHS/United States
- R29 CA070297/CA/NCI NIH HHS/United States
- R01 DK061002-05/DK/NIDDK NIH HHS/United States
- R56 DK061002/DK/NIDDK NIH HHS/United States
- R01 CA087767-01A2/CA/NCI NIH HHS/United States
- R01 CA087767/CA/NCI NIH HHS/United States
- R01 DK061002/DK/NIDDK NIH HHS/United States
- R01 CA070297-12/CA/NCI NIH HHS/United States
- R01 DK061002-02/DK/NIDDK NIH HHS/United States
- R01 CA070297-09/CA/NCI NIH HHS/United States
- CA 70297/CA/NCI NIH HHS/United States
- R01 CA070297-10/CA/NCI NIH HHS/United States
- R01 CA070297-06A1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases