Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced dna damage depends on functional TFIIH - PubMed (original) (raw)
Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced dna damage depends on functional TFIIH
Angelika Zotter et al. Mol Cell Biol. 2006 Dec.
Abstract
The structure-specific endonuclease XPG is an indispensable core protein of the nucleotide excision repair (NER) machinery. XPG cleaves the DNA strand at the 3' side of the DNA damage. XPG binding stabilizes the NER preincision complex and is essential for the 5' incision by the ERCC1/XPF endonuclease. We have studied the dynamic role of XPG in its different cellular functions in living cells. We have created mammalian cell lines that lack functional endogenous XPG and stably express enhanced green fluorescent protein (eGFP)-tagged XPG. Life cell imaging shows that in undamaged cells XPG-eGFP is uniformly distributed throughout the cell nucleus, diffuses freely, and is not stably associated with other nuclear proteins. XPG is recruited to UV-damaged DNA with a half-life of 200 s and is bound for 4 min in NER complexes. Recruitment requires functional TFIIH, although some TFIIH mutants allow slow XPG recruitment. Remarkably, binding of XPG to damaged DNA does not require the DDB2 protein, which is thought to enhance damage recognition by NER factor XPC. Together, our data present a comprehensive view of the in vivo behavior of a protein that is involved in a complex chromatin-associated process.
Figures
FIG. 1.
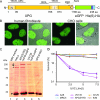
Expression and functionality of XPG-eGFP. (A) Schematic representation of the XPG-eGFP-His6-hemagglutinin (HA) fusion gene, with the N-terminal and C-terminal nuclease domains (N and C, respectively) and different interaction domains indicated. I, internal domain; PBD, PCNA binding domain; NLS, probable nuclear localization signal; aa, amino acids. (B) Localization of the XPG fusion protein in human fibroblasts (two images at left, showing the fluorescence signal and an overlay of fluorescence and phase contrast) and CHO cells (two images at right). XPG-eGFP is present mainly in the nucleus, except in the nucleoli. (C) Immunoblot (probed with monoclonal anti-XPG) of 40 μg of whole-cell extract from HeLa cells (lane 2), human XPCS1RO-Sv (XP-G) cells expressing XPG-eGFP (lane 3), CHO (UV135) cells expressing XPG-eGFP (lane 4), and untransfected XPCS1RO-Sv cells (lane 5). The molecular masses of protein markers are indicated in kilodaltons (kDa). eGFP-tagged XPG migrates slower than endogenous XPG (upper and lower arrows, respectively). No XPG protein was detected in the human fibroblasts, in which the severely truncated XPG-mRNA was probably highly unstable or not recognized. Chinese hamster ovary cell XPG was not detected with our anti-XPG serum. Loading control: PCNA (arrowhead). The asterisk indicates a cross-reacting nonspecific band only present in human cell extracts. (D) UV survival of repair-proficient human MRC5 cells (wild type; light blue line), XPCS1RO cells (violet line), XPCS1RO cells stably expressing XPG-eGFP (clone 5 cM; brown line), wild-type CHO cells (AA8; dark blue line), XPG-deficient CHO cells (UV135; purple line), and UV135 cells expressing XPG-eGFP (clone 129; yellow line). The transfected cell lines show a correction of UV sensitivity to the wild-type level.
FIG. 2.
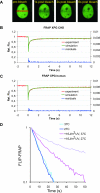
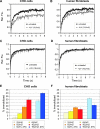
FRAP analysis of XPG mobility. (A) Example of FRAP analysis to determine effective diffusion coefficients in non-UV-irradiated cells. A strip (red rectangle) spanning the nucleus containing eGFP-tagged protein was bleached at high laser intensity. Subsequently, fluorescence recovery after photobleaching was monitored in the strip. (B and C) Graphical representation of FRAP analysis of eGFP-XPG in non-UV-irradiated CHO cells (B) and human fibroblasts (C). The mean relative fluorescence (Rel. Flu.) values (fluorescence after bleaching/fluorescence before bleaching) are plotted against the indicated times in seconds. Red lines, experimental data; green lines, simulated curves. Blue lines at the bottom of each graph represent residuals, which are a measure for the quality of the fits. (D) Simultaneous FLIP/FRAP analysis of XPG mobility in the nucleus of CHO cells. A small area at one pole of the nucleus was bleached for 1 s; subsequently, fluorescence was monitored over time in the bleached area (FRAP) and unbleached area (FLIP). The differences in eGFP intensity between the two areas after the bleach pulse are plotted on a log scale as a function of time. Light blue line, XPG redistribution at 37°C; dark blue line, XPG redistribution at 27°C; purple line, XPG redistribution in UV-irradiated cells at 37°C; violet line, XPG redistribution in UV-irradiated cells at 27°C. Experiments using UV-irradiated cells were performed between 10 and 30 min after global UV-C irradiation.
FIG. 3.
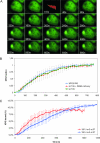
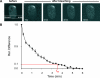
Accumulation of XPG-eGFP after local UV-DNA damage. (A) Time-lapse image series of XPG-eGFP expressed in CHO UV135 cells prior to and immediately after UV-C irradiation (100 J/m2). After preirradiation images were taken, cells were irradiated for 39 s (lightning arrow); subsequently, images were taken at 20 s intervals. (B) Incorporation kinetics of XPG-eGFP in CHO cells (UV135, green line; n = 5), CHO cells transfected with DDB2-mCherry (UV135 + DDB2, red line; n = 5), and human fibroblasts (XPCS1RO, blue line; n = 5) at UV-damaged areas after treatment with 100 J/m2 UV-C. The local relative accumulation of XPG-eGFP was measured versus time. (C) Incorporation kinetics of XPG-eGFP in UV135 cells at 37°C (red line; n = 5) and 27°C (blue line; n = 5). The local accumulation of XPG-eGFP was measured and plotted as a percentage of the total eGFP fluorescence of the cell nucleus (37°C, n = 11; 27°C, n = 20) versus time after the start of UV irradiation. Error bars represent standard deviations for the results of different experiments.
FIG. 4.
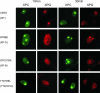
Accumulation of XPC and XPG after local UV damage in human wild-type (WT) cells (C5RO) and various TFIIH mutants at 10 min (columns 1 and 2) and 30 min (columns 3 and 4) after UV irradiation. Columns 1 and 3, immunofluorescence labeling with anti-XPC antibody (green); columns 2 and 4, labeling with anti-XPG antibody (red).
FIG. 5.
FRAP analysis of UV-treated and untreated cells to visualize DNA damage-dependent immobilization of XPG-eGFP. (A and B) FRAP recovery curves (normalized to prebleach intensity set to 1) for CHO cells and human fibroblasts, respectively. Black curves, XPG-eGFP recovery in untreated cells (as a reference); gray curves, recovery in UV-irradiated cells. Rel. Flu., relative fluorescence. (C and D) FRAP recovery curves (normalized to maximum recovery postbleach, which is set to 1) of XPG-eGFP in CHO cells and human fibroblasts, respectively. Black curves, XPG recovery in untreated cells (as a reference); gray curves, recovery in UV-irradiated cells. (E and F) Immobilized XPG-eGFP fractions in CHO cells and human fibroblasts, respectively, in response to different UV doses. Light blue and dark blue bars depict measurements of cells cultured at 27°C after treatment with 8 and 16 J/m2 UV, respectively. Error bars show the standard errors of the means.
FIG. 6.
FLIP analysis of locally UV-damaged areas in the nucleus. (A) A strip opposite a locally damaged area in the nucleus was bleached, and the redistribution of bleached and unbleached XPG-eGFP was monitored over time. (B) FLIP curve of the locally damaged nucleus. The relative differences between redistribution in the damaged area and that in the nondamaged area are shown plotted over time. Error bars depict the standard errors of the means.
Similar articles
- Novel functional interactions between nucleotide excision DNA repair proteins influencing the enzymatic activities of TFIIH, XPG, and ERCC1-XPF.
Winkler GS, Sugasawa K, Eker AP, de Laat WL, Hoeijmakers JH. Winkler GS, et al. Biochemistry. 2001 Jan 9;40(1):160-5. doi: 10.1021/bi002021b. Biochemistry. 2001. PMID: 11141066 - Definition of a short region of XPG necessary for TFIIH interaction and stable recruitment to sites of UV damage.
Thorel F, Constantinou A, Dunand-Sauthier I, Nouspikel T, Lalle P, Raams A, Jaspers NG, Vermeulen W, Shivji MK, Wood RD, Clarkson SG. Thorel F, et al. Mol Cell Biol. 2004 Dec;24(24):10670-80. doi: 10.1128/MCB.24.24.10670-10680.2004. Mol Cell Biol. 2004. PMID: 15572672 Free PMC article. - Regulation of endonuclease activity in human nucleotide excision repair.
Fagbemi AF, Orelli B, Schärer OD. Fagbemi AF, et al. DNA Repair (Amst). 2011 Jul 15;10(7):722-9. doi: 10.1016/j.dnarep.2011.04.022. Epub 2011 May 17. DNA Repair (Amst). 2011. PMID: 21592868 Free PMC article. Review. - Hot topics in DNA repair: the molecular basis for different disease states caused by mutations in TFIIH and XPG.
Schärer OD. Schärer OD. DNA Repair (Amst). 2008 Feb 1;7(2):339-44. doi: 10.1016/j.dnarep.2007.10.007. DNA Repair (Amst). 2008. PMID: 18077223 Free PMC article. Review.
Cited by
- PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1.
Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, Hensbergen P, Deelder A, de Groot A, Matsumoto S, Sugasawa K, Thoma N, Vermeulen W, Vrieling H, Mullenders L. Pines A, et al. J Cell Biol. 2012 Oct 15;199(2):235-49. doi: 10.1083/jcb.201112132. Epub 2012 Oct 8. J Cell Biol. 2012. PMID: 23045548 Free PMC article. - Chromatin structure and DNA damage repair.
Dinant C, Houtsmuller AB, Vermeulen W. Dinant C, et al. Epigenetics Chromatin. 2008 Nov 12;1(1):9. doi: 10.1186/1756-8935-1-9. Epigenetics Chromatin. 2008. PMID: 19014481 Free PMC article. - Generation of cell-based systems to visualize chromosome damage and translocations in living cells.
Roukos V, Burgess RC, Misteli T. Roukos V, et al. Nat Protoc. 2014 Oct;9(10):2476-92. doi: 10.1038/nprot.2014.167. Epub 2014 Sep 25. Nat Protoc. 2014. PMID: 25255091 Free PMC article. - Exploring new potential role of DDB2 by host cell reactivation assay in human tumorigenic cells.
Bassi E, Perucca P, Guardamagna I, Prosperi E, Stivala LA, Cazzalini O. Bassi E, et al. BMC Cancer. 2019 Oct 29;19(1):1013. doi: 10.1186/s12885-019-6258-0. BMC Cancer. 2019. PMID: 31664956 Free PMC article.
References
- Botta, E., T. Nardo, A. R. Lehmann, J. M. Egly, A. M. Pedrini, and M. Stefanini. 2002. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet. 11:2919-2928. - PubMed
- de Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768-785. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous