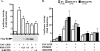A novel cyclic AMP-dependent Epac-Rit signaling pathway contributes to PACAP38-mediated neuronal differentiation - PubMed (original) (raw)
A novel cyclic AMP-dependent Epac-Rit signaling pathway contributes to PACAP38-mediated neuronal differentiation
Geng-Xian Shi et al. Mol Cell Biol. 2006 Dec.
Abstract
Pituitary adenylate cyclase-activating polypeptide (PACAP38) stimulation results in the activation of G(s)alpha protein-coupled receptors to regulate neuronal differentiation in a cyclic AMP (cAMP)-dependent manner. These pathways involve protein kinase A (PKA)-dependent processes, but a growing body of evidence indicates that cAMP also regulates cellular functions through PKA-independent signaling cascades. Here we show that the Rit small GTPase is regulated by PACAP38 in a cAMP-dependent but PKA-independent fashion. Rit activation results from stimulation of the cAMP-activated guanine nucleotide exchange factor Epac but does not appear to rely upon the activation of Rap GTPases, the accepted cellular Epac substrates. Although RNA interference studies demonstrated that Epac is required for PACAP38-mediated Rit activation, neither Epac1 nor Epac2 activates Rit directly, indicating that Epac signals to Rit through a novel mechanism in which Rap signaling is not essential. Loss-of-function analysis demonstrated that Rit makes an important contribution to PACAP38-mediated neuronal differentiation. Surprisingly, although Rit is required for sustained extracellular signal-regulated kinase (ERK) and p38 mitogen-activated protein kinase signaling following nerve growth factor stimulation of pheochromocytoma 6 (PC6) cells, Rit silencing selectively suppressed PACAP38-elicited activation of p38, without obvious effects on ERK signaling in the same cells. Moreover, the ability of PACAP38 to stimulate CREB-dependent transcription and to promote neurite outgrowth was inhibited by Rit knockdown. Together, these studies identify an unsuspected connection between cAMP and Rit signaling pathways and imply that Rit can function downstream of G(s)alpha/cAMP/Epac in a novel signal transduction pathway necessary for PACAP38-mediated neuronal differentiation and CREB signaling.
Figures
FIG. 1.
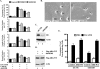
Rit is required for PACAP38-mediated neuronal differentiation. (A and B) Rit knockdown attenuates PACAP-induced neurite outgrowth. PC6 cells were transfected with either a control shRNA vector (shCTR) or a Rit-selective shRNA (shRit208), neurite formation was induced with PACAP38 (5 nM), and transformed cells were enriched by drug selection (G418). On days 3 and 7 after PACAP38 stimulation, the percentages of neurite-bearing cells, neurite numbers, neurite lengths, and neurite branches were counted or calculated as described in Materials and Methods. The results are shown for day 7 as means ± standard deviations (SD [error bars]) for three independent experiments. Pretreatment of control cells with either a MEK/ERK (10 μM PD98059) or p38 (10 μM SB203580) kinase inhibitor was used as a control. Representative micrographs from 7-day cultures are shown (B). (C) shRit208 mediates Rit silencing. PC6 cells were transfected with 1.5 μg of shRit208 and subjected to G418 selection (400 μg/ml, 48 h). Endogenous Rit protein levels were determined by immunoblot analysis with an anti-Rit monoclonal antibody. (D) Human Rit is not subject to shRit208-mediated silencing. PC6 cells were transfected with 0.2 μg of 3× Flag-hRit-WT in the presence of 1.5 μg of shRit208 and subjected to G418 (400 μg/ml) selection for 48 h. The expression levels of 3× Flag-hRit-WT were determined by anti-Flag immunoblotting, and actin expression served as a loading control. (E) Human Rit (hRit) restores PACAP38-induced neurite outgrowth to shRit208-expressing PC6 cells. PC6 cells were transfected as described for panel D, and differentiation was stimulated with PACAP38 as described for panel A. Neurite outgrowth was analyzed on day 3 after PACAP38 stimulation, and the percentages of neurite-bearing cells are shown as means ± SD for three independent experiments.
FIG. 2.
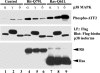
Rit is required for PACAP38-mediated p38 activation. (A) Rit silencing attenuates PACAP38-mediated p38 MAP kinase signaling but is not required for ERK activation. PC6 cells were transfected with shCTR or shRit208 and enriched by G418 (400 μg/ml) selection for 2 days. Cells were then serum starved for 5 h prior to PACAP38 (5 nM) stimulation, and the phosphorylation status of ERK1/2 and p38 MAP kinases was analyzed by immunoblotting with the appropriate phospho-specific antibodies. (B) Human Rit restores PACAP38-mediated p38 signaling to shRit208-expressing PC6 cells. PC6 cells were transfected as described in the legend to Fig. 1D and subjected to G418 (400 μg/ml) selection for 48 h. Prior to the preparation of whole-cell lysates, cells were serum starved for 5 h and stimulated with PACAP38 (10 nM) for 15 min, as indicated. The levels of phosphorylated p38 and ERK1/2 MAP kinases were analyzed by immunoblotting with phospho-specific antibodies. The expression levels of human Rit and actin levels were examined by Western blotting. The results shown are from one experiment that was representative of the four experiments performed.
FIG. 3.
Rit activates p38α and p38γ MAP kinases. PC6 cells were cotransfected with Flag-tagged p38 MAP kinase isoforms α, γ, and δ in the presence of empty 3× HA vector (control), 3× HA-hRit-Q79L, or 3× HA-H-Ras-Q61L. The transfected p38 isoforms were immunoprecipitated (I.P.) with an anti-Flag monoclonal antibody from 400 μg of total cell lysate, and the immunoprecipitates were subjected to in vitro p38 MAP kinase assays as described in Materials and Methods. The data shown are representative of three independent experiments.
FIG. 4.
Rit is involved in PACAP38-mediated CREB activation. (A) PC6 cells were transfected with 100 ng of pFA2-CREB, pFR-Luciferase reporter, and pRSV-β-gal in the absence or presence of 3× Flag-Rit-S35N (0.25, 0.5, and 1.0 μg) or shRit208, as indicated. Cells were serum starved for 5 h prior to PACAP38 (10 nM) stimulation for 4 h where indicated, and the luciferase activities were analyzed as described in Materials and Methods. The results are presented as means ± SD for three experiments performed in duplicate. (B) p38 MAP kinase signaling contributes to PACAP38/Rit signaling to CREB. PC6 cells were transfected with the CREB reporter system as described for panel A in the presence of either empty 3× Flag vector (EV), 3× Flag-RitWT, or 3× Flag-Rit-Q79L. The cells were serum starved for 5 h and pretreated with either PD98059, SB203580, or a combination of both (10 μM each) for 30 min before PACAP38 stimulation (10 nM for 4 h). The results are presented as means ± SD for 4 to 10 experiments performed in triplicate.
FIG. 5.
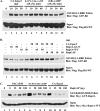
PACAP38 activates Rit in a Gsα-dependent manner. (A) PACAP38 activates Rit in PC6 cells. PC6 cells were transfected with an expression construct for 3× Flag-hRit-WT. Prior to preparation of whole-cell lysates, cells were serum starved for 5 h and then exposed to 10 nM PACAP38 for the indicated time (lanes 2 to 7) or to the indicated concentrations of PACAP38 for 15 min (lanes 8 to 12). Whole-cell lysates (200 μg) were subjected to GST pull-down assay, using GST-RGL3-RBD agarose, and the levels of GTP-bound Rit were determined as described in Materials and Methods. Total lysates (5 μg) were immunoblotted with an anti-Flag monoclonal antibody to ensure equal expression of 3× Flag-hRit-WT in the assay. (B) Gsα and CTX, but not Gβγ, activate Rit. PC6 cells were cotransfected with expression constructs for 3× Flag-hRit-WT and either a constitutively active Gsα (GsαQL) mutant or a combination of Gβ1 and Gγ2 vectors, as indicated, and Rit activation was determined using a GST-RGL3-RBD pull-down assay as described above. To examine the requirement for adenylate cyclase/cAMP in Rit activation, serum-starved (5 h) PC6 cells transfected with 3× Flag-Rit-WT were treated with or without ddA (50 μM) for 5 min and then exposed to CTX (5 μg/ml) for the indicated time periods, and the levels of GTP-bound Rit were determined using GST-RGL3-RBD as described above.
FIG. 6.
PACAP38-mediated Rit activation is cAMP dependent but PKA independent. PC6 cells expressing 3× Flag-hRit-WT were preincubated with H89 (10 μM, 60 min) or ddA (50 μM, 5 min) and then stimulated with PACAP38 (10 nM) or exposed to 8-Br-cAMP (50 μM) for the indicated times. Cell lysates (200 μg) were analyzed by GST-RGL3-RBD pull-down assay as described in Materials and Methods, and GTP-bound Rit levels were determined by immunoblot analysis using biotinylated anti-Flag antibody. Equal expression of recombinant Flag-tagged human Rit was controlled by immunoblotting cell lysates (5 μg) with anti-Flag antibody (input).
FIG. 7.
Epac induces the activation of Rit in PC6 cells. (A) Epac2 induces Rit activation in PC6 cells. PC6 cells were transfected with 3× Flag-hRit-WT (1 μg), with or without constitutively active Epac2 (10, 20, or 50 ng). Prior to the preparation of whole-cell lysates, cells were serum starved for 5 h. Rit-GTP was recovered using GST-RGL3-RBD pull-down analysis as described in Materials and Methods. PC6 cells transfected with 3× Flag-hRit-WT were exposed to either 8-Br-cAMP (25 μM) or the Epac-selective cAMP analog 8-CPT-2′-_O_-Me-cAMP (25 μM) for the indicated times, and the levels of GTP-bound Rit were analyzed using a GST-RGL3-RBD pull-down assay. (B) Epac-mediated Rit activation does not appear to require Rap1A. PC6 cells were cotransfected with 3× Flag-hRit-WT and an active form of Rap1A (Rap1A-G12V), wild-type Rap1A (Rap1A-WT), or RapGAP in the presence or absence of constitutively active Epac2 (CA-Epac2), as indicated. Transfected cells were serum starved for 5 h, whole-cell lysates were prepared, and Rit activation was accessed using GST-RGL3-RBD pull-down analysis. The expression level of recombinant Rit was determined by immunoblot analysis. The data shown are from one experiment that is representative of the four experiments performed. (C) RapGAP inhibits Rap1A activation. PC6 cells expressing Myc-Rap1A-WT in the presence of Flag-RapGAP or empty Flag vector were serum starved and then stimulated with PACAP38 (10 nM, 10 min) (lanes 2 to 5) or 8-CPT-2-Me-cAMP (25 μM, 10 min) (lanes 6 to 9) or cotransfected with Flag-CA-Epac2 (50 ng) (lanes 10 to 13). The GTP-loading levels of Rap1A were determined as described in Materials and Methods.
FIG. 8.
Epac proteins are not direct RitGEFs. The graphs show the nucleotide exchange of Rit. The dissociation of the Rit-mGDP complex was monitored by the time-dependent decrease in fluorescence intensity. Rit-mGDP (200 nM) was loaded with the fluorescent nucleotide analogue mGDP in the presence (squares) or absence (circles) of inactive (open squares) or activated (500 μM cAMP treated) (closed squares) 100 nM Epac1 (left panel) or 100 nM Epac2 (right panel) after the addition of a 100-fold excess of unlabeled GDP, as indicated. For clarity of presentation, the curves were shifted 0.025 relative units to avoid the overlay of symbols.
FIG. 9.
Epac1 is required for Rit activation in PC6 cells. (A) Expression of shEpac1-1501 induces Epac1 silencing. PC6 cells were transfected with shCTR (lane 2), shEpac1-1501 (lane 3), shEpac1-2000 (lane 4), or shEpac2-979 (lane 5) and subjected to G418 (400 μg/ml) enrichment for 2 days. Purified RNAs were isolated and subjected to RT-PCR using Epac1-, Epac2-, and actin-specific primers. Non-reverse transcription controls are shown (lane 1). M, 1-kb Plus DNA ladder (Invitrogen). (B) Epac1-1501 silencing attenuates PACAP38-dependent Rit activation. PC6 cells were cotransfected with 3× Flag-Rit-WT and either shCTR, shEpac1-1501, shEpac1-2000, or shEpac2-979. The cells were starved with serum-free DMEM for 5 h and stimulated with 10 nM PACAP38, and the level of GTP-bound Rit was determined as described in Materials and Methods. (C) Epac1 and Epac2 restore cAMP-mediated Rit activation to shEpac1-1501-expressing PC6 cells. PC6 cells were transfected with 3× Flag-Rit-WT in the presence of shCTR or shEpac1-1501, together with an empty 3× Flag, WT Epac1, or WT Epac2 expression vector, and stimulated with 8-CPT-2′-_O_-Me-cAMP (25 μM) for the indicated times. The GTP-Rit levels were analyzed using a GST-RGL3-RBD pull-down assay as described above.
Similar articles
- Pituitary adenylate cyclase-activating polypeptide 38-mediated Rin activation requires Src and contributes to the regulation of HSP27 signaling during neuronal differentiation.
Shi GX, Jin L, Andres DA. Shi GX, et al. Mol Cell Biol. 2008 Aug;28(16):4940-51. doi: 10.1128/MCB.02193-07. Epub 2008 Jun 9. Mol Cell Biol. 2008. PMID: 18541665 Free PMC article. - Src-dependent TrkA transactivation is required for pituitary adenylate cyclase-activating polypeptide 38-mediated Rit activation and neuronal differentiation.
Shi GX, Jin L, Andres DA. Shi GX, et al. Mol Biol Cell. 2010 May 1;21(9):1597-608. doi: 10.1091/mbc.e09-12-1033. Epub 2010 Mar 10. Mol Biol Cell. 2010. PMID: 20219970 Free PMC article. - Rit subfamily small GTPases: regulators in neuronal differentiation and survival.
Shi GX, Cai W, Andres DA. Shi GX, et al. Cell Signal. 2013 Oct;25(10):2060-8. doi: 10.1016/j.cellsig.2013.06.002. Epub 2013 Jun 11. Cell Signal. 2013. PMID: 23770287 Free PMC article. Review. - Cell physiology of cAMP sensor Epac.
Holz GG, Kang G, Harbeck M, Roe MW, Chepurny OG. Holz GG, et al. J Physiol. 2006 Nov 15;577(Pt 1):5-15. doi: 10.1113/jphysiol.2006.119644. Epub 2006 Sep 14. J Physiol. 2006. PMID: 16973695 Free PMC article. Review.
Cited by
- Adenosine 2A receptor promotes collagen production by human fibroblasts via pathways involving cyclic AMP and AKT but independent of Smad2/3.
Perez-Aso M, Fernandez P, Mediero A, Chan ES, Cronstein BN. Perez-Aso M, et al. FASEB J. 2014 Feb;28(2):802-12. doi: 10.1096/fj.13-241646. Epub 2013 Nov 7. FASEB J. 2014. PMID: 24200882 Free PMC article. - Claulansine F promotes neuritogenesis in PC12 cells via the ERK signaling pathway.
Ma YZ, Ning N, He WB, Li JW, Hu JF, Chu SF, Chen NH. Ma YZ, et al. Acta Pharmacol Sin. 2013 Dec;34(12):1499-507. doi: 10.1038/aps.2013.95. Epub 2013 Oct 7. Acta Pharmacol Sin. 2013. PMID: 24096602 Free PMC article. - Neuroprotective role of prostaglandin PGE2 EP2 receptor in hemin-mediated toxicity.
Mohan S, Narumiya S, Doré S. Mohan S, et al. Neurotoxicology. 2015 Jan;46:53-9. doi: 10.1016/j.neuro.2014.10.012. Epub 2014 Nov 13. Neurotoxicology. 2015. PMID: 25451967 Free PMC article. - Rit signaling contributes to interferon-gamma-induced dendritic retraction via p38 mitogen-activated protein kinase activation.
Andres DA, Shi GX, Bruun D, Barnhart C, Lein PJ. Andres DA, et al. J Neurochem. 2008 Dec;107(5):1436-47. doi: 10.1111/j.1471-4159.2008.05708.x. Epub 2008 Oct 24. J Neurochem. 2008. PMID: 18957053 Free PMC article. - PKA and Epac synergistically inhibit smooth muscle cell proliferation.
Hewer RC, Sala-Newby GB, Wu YJ, Newby AC, Bond M. Hewer RC, et al. J Mol Cell Cardiol. 2011 Jan;50(1):87-98. doi: 10.1016/j.yjmcc.2010.10.010. Epub 2010 Oct 30. J Mol Cell Cardiol. 2011. PMID: 20971121 Free PMC article.
References
- Barrie, A. P., A. M. Clohessy, C. S. Buensuceso, M. V. Rogers, and J. M. Allen. 1997. Pituitary adenylyl cyclase-activating peptide stimulates extracellular signal-regulated kinase 1 or 2 (ERK1/2) activity in a Ras-independent, mitogen-activated protein kinase/ERK kinase 1 or 2-dependent manner in PC12 cells. J. Biol. Chem. 272:19666-19671. - PubMed
- Bito, H., and S. Takemoto-Kimura. 2003. Ca(2+)/CREB/CBP-dependent gene regulation: a shared mechanism critical in long-term synaptic plasticity and neuronal survival. Cell Calcium 34:425-430. - PubMed
- Bos, J. L. 2003. Epac: a new cAMP target and new avenues in cAMP research. Nat. Rev. Mol. Cell Biol. 4:733-738. - PubMed
- Bos, J. L., J. de Rooij, and K. A. Reedquist. 2001. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2:369-377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 RR 20171/RR/NCRR NIH HHS/United States
- P20 RR020171/RR/NCRR NIH HHS/United States
- R01 NS045103/NS/NINDS NIH HHS/United States
- R56 NS045103/NS/NINDS NIH HHS/United States
- NS 045103/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous