Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus - PubMed (original) (raw)
. 2006 Oct 5;443(7111):578-81.
doi: 10.1038/nature05181. Epub 2006 Sep 27.
Terrence M Tumpey, Sean C Proll, Victoria Carter, Olivia Perwitasari, Matthew J Thomas, Christopher F Basler, Peter Palese, Jeffery K Taubenberger, Adolfo García-Sastre, David E Swayne, Michael G Katze
Affiliations
- PMID: 17006449
- PMCID: PMC2615558
- DOI: 10.1038/nature05181
Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus
John C Kash et al. Nature. 2006.
Abstract
The influenza pandemic of 1918-19 was responsible for about 50 million deaths worldwide. Modern histopathological analysis of autopsy samples from human influenza cases from 1918 revealed significant damage to the lungs with acute, focal bronchitis and alveolitis associated with massive pulmonary oedema, haemorrhage and rapid destruction of the respiratory epithelium. The contribution of the host immune response leading to this severe pathology remains largely unknown. Here we show, in a comprehensive analysis of the global host response induced by the 1918 influenza virus, that mice infected with the reconstructed 1918 influenza virus displayed an increased and accelerated activation of host immune response genes associated with severe pulmonary pathology. We found that mice infected with a virus containing all eight genes from the pandemic virus showed marked activation of pro-inflammatory and cell-death pathways by 24 h after infection that remained unabated until death on day 5. This was in contrast with smaller host immune responses as measured at the genomic level, accompanied by less severe disease pathology and delays in death in mice infected with influenza viruses containing only subsets of 1918 genes. The results indicate a cooperative interaction between the 1918 influenza genes and show that study of the virulence of the 1918 influenza virus requires the use of the fully reconstructed virus. With recent concerns about the introduction of highly pathogenic avian influenza viruses into humans and their potential to cause a worldwide pandemic with disastrous health and economic consequences, a comprehensive understanding of the global host response to the 1918 virus is crucial. Moreover, understanding the contribution of host immune responses to virulent influenza virus infections is an important starting point for the identification of prognostic indicators and the development of novel antiviral therapies.
Figures
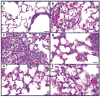
Figure 1. Photomicrographs of lung sections from mice infected with influenza A virus, stained with haematoxylin and eosin
a, Mild histiocytic alveolitis with normal bronchiole, Tx/91 virus, day 3. b, Moderate diffuse alveolitis, predominantly neutrophilic, but with macrophages and associated haemorrhage and oedema, r1918 virus, day 3. c, Severe focal alveolitis with neutrophils distending affected alveoli, r1918 virus, day 3. d, e, Moderate histiocytic alveolitis and some neutrophils and oedema, 5:3 1918 (d) and 2:6 1918 (e) viruses, day 3. f, Severe diffuse histiocytic alveolitis with severe oedema, r1918 virus, day 5. Scale bar, 20μm.
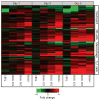
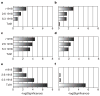
Figure 2. Increased and earlier expression of genes associated with the activation of key immune cells in mouse lung infected with r1918 influenza virus
Expression of selected immune-cell-related genes in the lungs of mice infected with Tx91, 2:6 1918, 5:3 1918 or r1918 viruses on days 1, 3 and 5 after infection, as determined by expression microarray analysis. For each infection point, the data presented are the error-weighted average expression changes calculated from four technical replicate arrays performed on three individual mice (n = 12 total). Genes shown in red were upregulated and genes shown in green were downregulated in infected relative to mock-infected mouse lung. NK, natural killer; TH1, T helper 1.
Figure 3. Increased activation of inflammatory and death receptor gene expression by r1918 influenza virus
a–d, Analysis of the biological response pathways that showed most significant activation during r1918 infection, including death receptor (a), interleukin-6 (b), interferon (c) and Toll-like receptor (d) pathways. e, f, The biological responses most significantly induced in the non-lethal Tx91 infections: G2/M checkpoint control (e) and glutathione metabolism (f). Pathway analysis was performed on genes that were induced more than twofold (P < 0.01) in any of the infections compared with uninfected controls. For this purpose, Ingenuity Pathway Analysis was used, which analyses the RNA expression data in the context of known biological response and regulatory networks, as well as other higher-order response pathways to assign functional information and biological relevance. These results are shown as the negative logarithm of significance, which is a statistical score and is a measure of the likelihood of the genes in a given network being found together as a result of chance, as determined by Fisher’s exact test (see Methods for details).
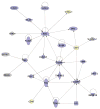
Figure 4. Functional relationships of activated of cell death responses during infection with r1918 influenza virus
Biological network of selected genes that were only induced more than twofold (P < 0.01) in the r1918-infected mouse lung compared with uninfected controls. This diagram shows the direct (solid lines) and indirect (dashed lines) interactions reported for these cell-death-related (blue shading) and immune-response-related (yellow shading) genes; grey denotes genes with multiple and/or undefined biological functions. Biological network analysis was performed by using Ingenuity Pathway Analysis and showed statistical significance with Fisher’s exact test to calculate a _P_-value determining the probability that each biological function and/or disease assigned to that data set was due to chance alone (see Methods for details).
Similar articles
- The 1918 Influenza Virus PB2 Protein Enhances Virulence through the Disruption of Inflammatory and Wnt-Mediated Signaling in Mice.
Forero A, Tisoncik-Go J, Watanabe T, Zhong G, Hatta M, Tchitchek N, Selinger C, Chang J, Barker K, Morrison J, Berndt JD, Moon RT, Josset L, Kawaoka Y, Katze MG. Forero A, et al. J Virol. 2015 Dec 9;90(5):2240-53. doi: 10.1128/JVI.02974-15. J Virol. 2015. PMID: 26656717 Free PMC article. - Implication of inflammatory macrophages, nuclear receptors, and interferon regulatory factors in increased virulence of pandemic 2009 H1N1 influenza A virus after host adaptation.
Josset L, Belser JA, Pantin-Jackwood MJ, Chang JH, Chang ST, Belisle SE, Tumpey TM, Katze MG. Josset L, et al. J Virol. 2012 Jul;86(13):7192-206. doi: 10.1128/JVI.00563-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532695 Free PMC article. - Differential pulmonary transcriptomic profiles in murine lungs infected with low and highly virulent influenza H3N2 viruses reveal dysregulation of TREM1 signaling, cytokines, and chemokines.
Ivan FX, Rajapakse JC, Welsch RE, Rozen SG, Narasaraju T, Xiong GM, Engelward BP, Chow VT. Ivan FX, et al. Funct Integr Genomics. 2012 Mar;12(1):105-17. doi: 10.1007/s10142-011-0247-y. Epub 2011 Aug 28. Funct Integr Genomics. 2012. PMID: 21874528 - Use of functional genomics to understand influenza-host interactions.
Fornek JL, Korth MJ, Katze MG. Fornek JL, et al. Adv Virus Res. 2007;70:81-100. doi: 10.1016/S0065-3527(07)70003-9. Adv Virus Res. 2007. PMID: 17765704 Free PMC article. Review. - Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses.
Korth MJ, Tchitchek N, Benecke AG, Katze MG. Korth MJ, et al. Semin Immunol. 2013 Oct 31;25(3):228-39. doi: 10.1016/j.smim.2012.11.001. Epub 2012 Dec 5. Semin Immunol. 2013. PMID: 23218769 Free PMC article. Review.
Cited by
- Mast cells and influenza a virus: association with allergic responses and beyond.
Graham AC, Temple RM, Obar JJ. Graham AC, et al. Front Immunol. 2015 May 18;6:238. doi: 10.3389/fimmu.2015.00238. eCollection 2015. Front Immunol. 2015. PMID: 26042121 Free PMC article. Review. - Kinetic characterization of PB1-F2-mediated immunopathology during highly pathogenic avian H5N1 influenza virus infection.
Leymarie O, Jouvion G, Hervé PL, Chevalier C, Lorin V, Lecardonnel J, Da Costa B, Delmas B, Escriou N, Le Goffic R. Leymarie O, et al. PLoS One. 2013;8(3):e57894. doi: 10.1371/journal.pone.0057894. Epub 2013 Mar 1. PLoS One. 2013. PMID: 23469251 Free PMC article. - The 1918 Influenza Pandemic and Its Legacy.
Taubenberger JK, Morens DM. Taubenberger JK, et al. Cold Spring Harb Perspect Med. 2020 Oct 1;10(10):a038695. doi: 10.1101/cshperspect.a038695. Cold Spring Harb Perspect Med. 2020. PMID: 31871232 Free PMC article. Review. - Embracing the heterogeneity of natural viruses in mouse studies.
Fiege JK, Langlois RA. Fiege JK, et al. J Gen Virol. 2022 Jun;103(6):001758. doi: 10.1099/jgv.0.001758. J Gen Virol. 2022. PMID: 35737518 Free PMC article. - Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection.
Cilloniz C, Pantin-Jackwood MJ, Ni C, Goodman AG, Peng X, Proll SC, Carter VS, Rosenzweig ER, Szretter KJ, Katz JM, Korth MJ, Swayne DE, Tumpey TM, Katze MG. Cilloniz C, et al. J Virol. 2010 Aug;84(15):7613-24. doi: 10.1128/JVI.00553-10. Epub 2010 May 26. J Virol. 2010. PMID: 20504916 Free PMC article.
References
- Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 ‘Spanish’ influenza pandemic. Bull Hist Med. 2002;76:105–115. - PubMed
- Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. - PubMed
- Kobasa D, et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources